Method and kit for directly amplifying DNA (deoxyribonucleic acid) bar code of trace sample without extraction
A very small, extraction-free technology, applied in the biological field, can solve the problems of inability to achieve accurate species identification, insufficient effective data, etc., and achieve the effects of high reliability and adaptability, high amplification efficiency and good stability.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0024] Example 1: DNA template amplified DNA barcode sequence prepared by different concentrations of lysate
[0025] Prepare 100 ml of NaOH solutions with concentrations of 10, 25, 50, 75, and 100 mM respectively, and autoclave them for later use.
[0026] Preparation of buffer: 1.22g Tris, 7.46g KCl, 0.3g EDTA plus deionized water to make up to 100ml, adjust the pH to 9.5, autoclave for later use.
[0027] Take 1 / 5 hind tarsus (about 10 μg) of five flies and put them into five 1.5ml centrifuge tubes, add liquid nitrogen, grind them into powder with a grinding rod, add 10 and 25 μg to the five centrifuge tubes respectively , 50, 75, and 100 mM NaOH solution 180 μl, placed at 95°C for 10 minutes, then briefly centrifuged at 4000 rpm for 5 seconds, then added 20 μl of buffer solution, placed at 95°C for 10 minutes, centrifuged at 12,000 rpm for 1 minute, discarded the precipitate, and the obtained supernatant was the fly DNA solution.
[0028] Using the fly DNA solution prepar...
Embodiment 2
[0032] Example 2: Preparation of DNA Template Amplified DNA Barcode Sequence by Mixing Buffer and Lysis Solution in Different Volume Ratio
[0033] The preparation method of lysate and buffer is the same as in Example 1.
[0034] Take 1 / 5 hind tarsus (about 10 μg) of five flies and put them into five 1.5ml centrifuge tubes respectively, add liquid nitrogen, grind them into powder with a grinding rod, and add 50 mM NaOH to the five centrifuge tubes respectively Solution 20, 60, 120, 180, 200μl, placed at 95°C for 10min, centrifuged briefly at 4000rpm for 5s, then added 180, 140, 80, 20, 0μl of buffer respectively, placed at 95°C for 10min, centrifuged at 12000rpm for 1min, discarded the precipitate, and obtained The supernatant is the fly DNA solution.
[0035] The PCR amplification reaction system and reaction conditions were the same as in Example 1.
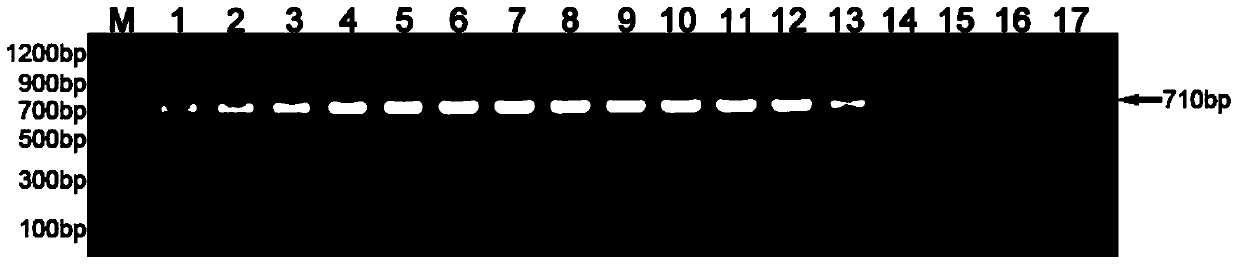
[0036] Amplification results such as figure 2 As shown, the negative control is ddH 2 O is the result of template amplif...
Embodiment 3
[0038] Example 3: Amplification of DNA barcode sequences at different annealing temperatures
[0039] The preparation method of lysate and buffer is the same as in Example 1.
[0040] Take 16 fly 1 / 5 hind tarsus (about 10μg) and put them into 16 1.5ml centrifuge tubes, add liquid nitrogen, grind them into powder with a grinding rod, and then add 50mM NaOH solution to the centrifuge tubes respectively 180 μl, placed at 95°C for 10 minutes, then briefly centrifuged at 4000rpm for 5s, then added 20μl of buffer solution, placed at 95°C for 10 minutes, centrifuged at 12000rpm for 1min, discarded the precipitate, and the obtained supernatant was the fly DNA solution.
[0041] The PCR amplification reaction system was the same as in Example 1.
[0042] The annealing temperatures of the 16 samples were 32, 34, 36, 38, 40, 42, 44, 46, 48, 50, 52, 54, 56, 58, 60, and 62°C, respectively.
[0043] Other amplification reaction conditions are the same as in Example 1.
[0044] Amplificat...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com