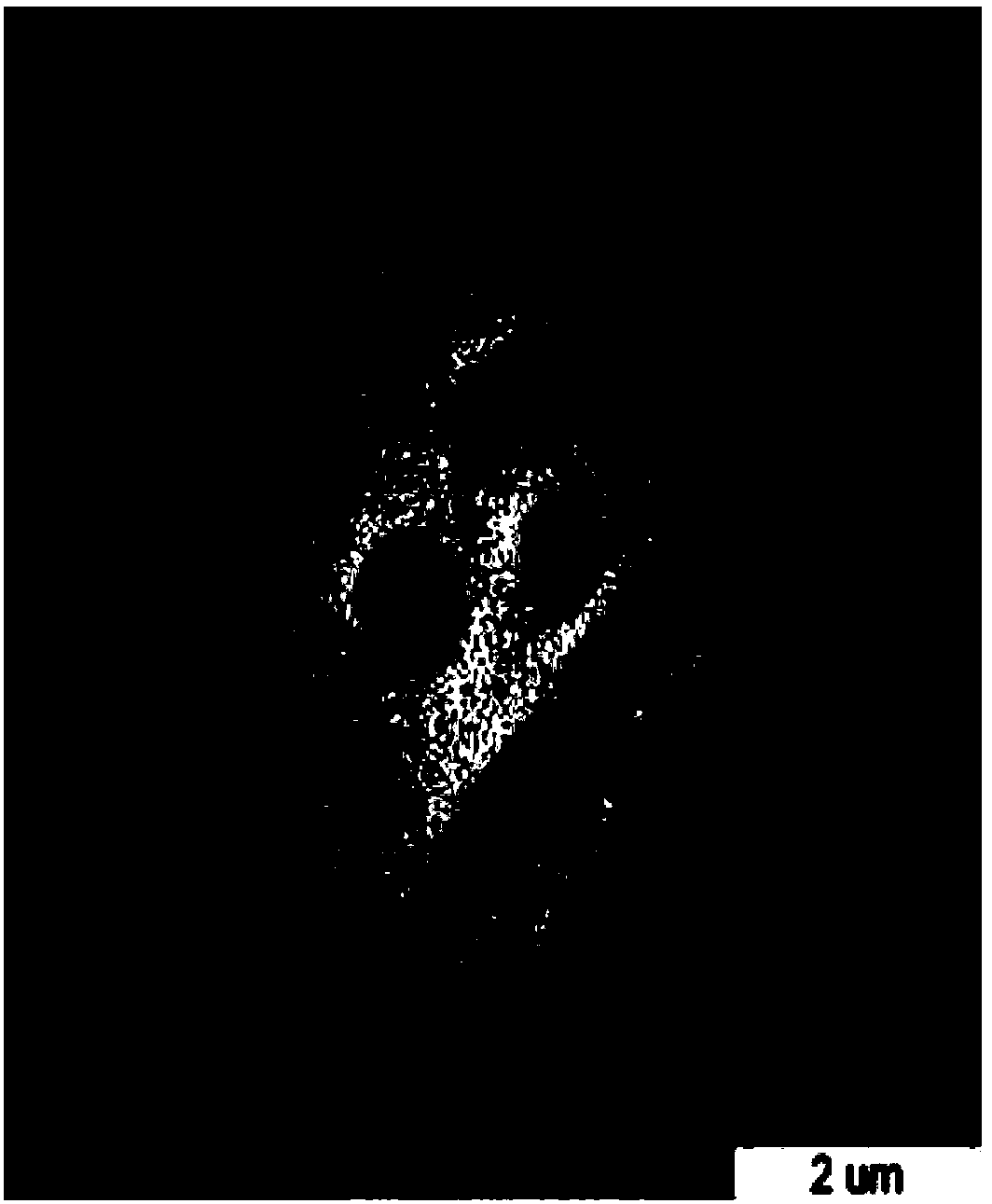Construction method of bacterial ghost carrier
A construction method and carrier technology, applied in the field of genetic engineering, can solve problems such as interfering with the expression of cleavage gene E
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
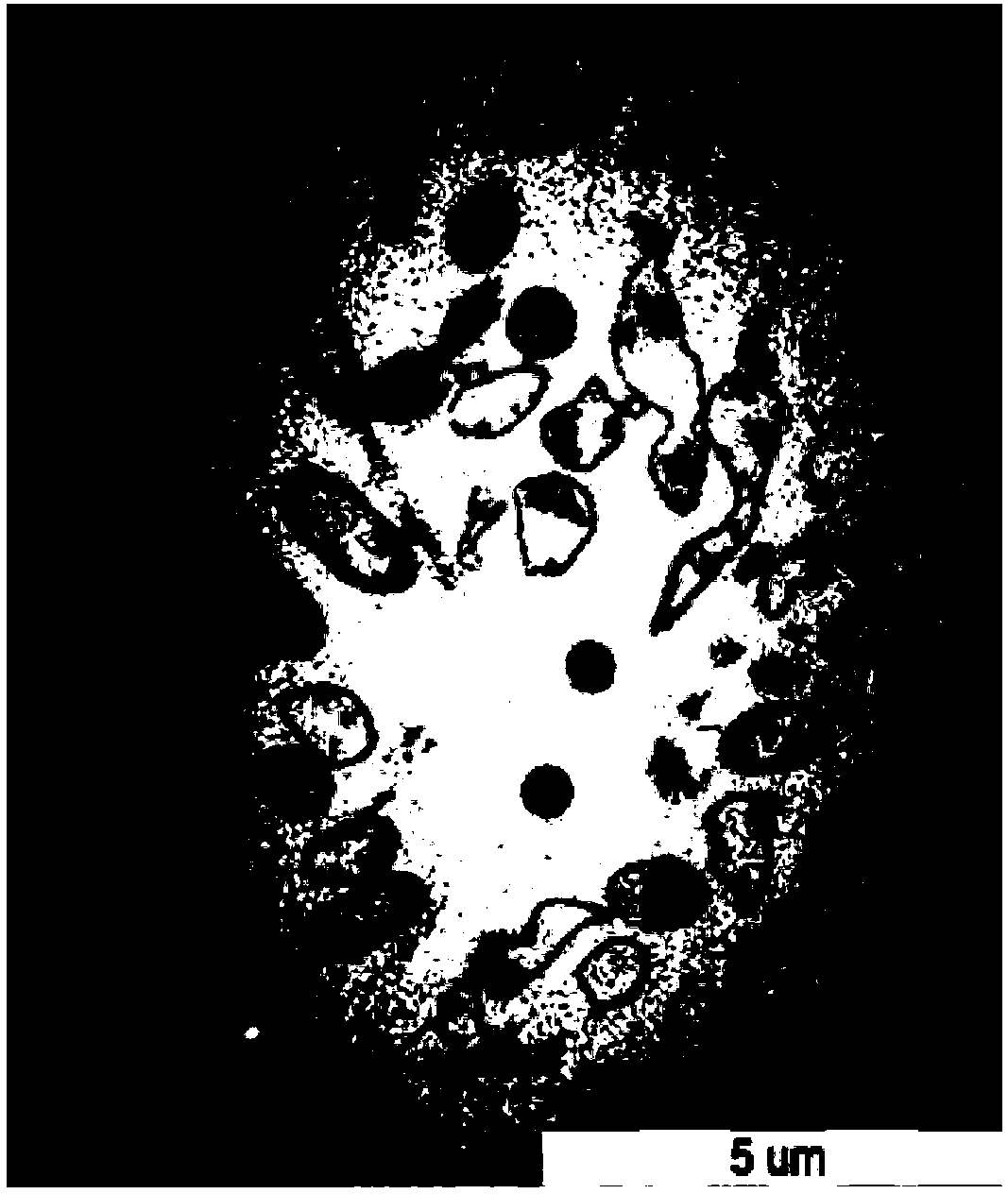
[0044] Example 1 Preparation of Escherichia coli ghosts with pBV220 as carrier
[0045] 1. Design and synthesis of E gene
[0046] Using Oligo6.0 and primer5.0 software, primers were designed according to the coding sequence of phage PhiX174 cleavage gene E (NC-001422) registered in GenBank. A restriction enzyme cutting site EcoRI was introduced at the 5' end of the upstream primer, a restriction enzyme cutting site Sal I restriction enzyme cutting site was introduced at the downstream 5' end, and several protective bases were randomly added at both ends. The primers were synthesized by Shanghai Sangong Synthesis Department.
[0047] E upstream primer E1: 5'-CGC GAATTCATGGTACGCTGGACTTTGT G-3', the sequence in the underline is the restriction enzyme cutting site EcoRI enzyme cutting site.
[0048] E downstream primer E2: 5′-ACG CGTCGA CTCACTCCTTCCGCACG TAAT-3', the underlined sequence is the restriction enzyme cutting site Sal I enzyme cutting site.
[0049] 2. PCR ampli...
Embodiment 2
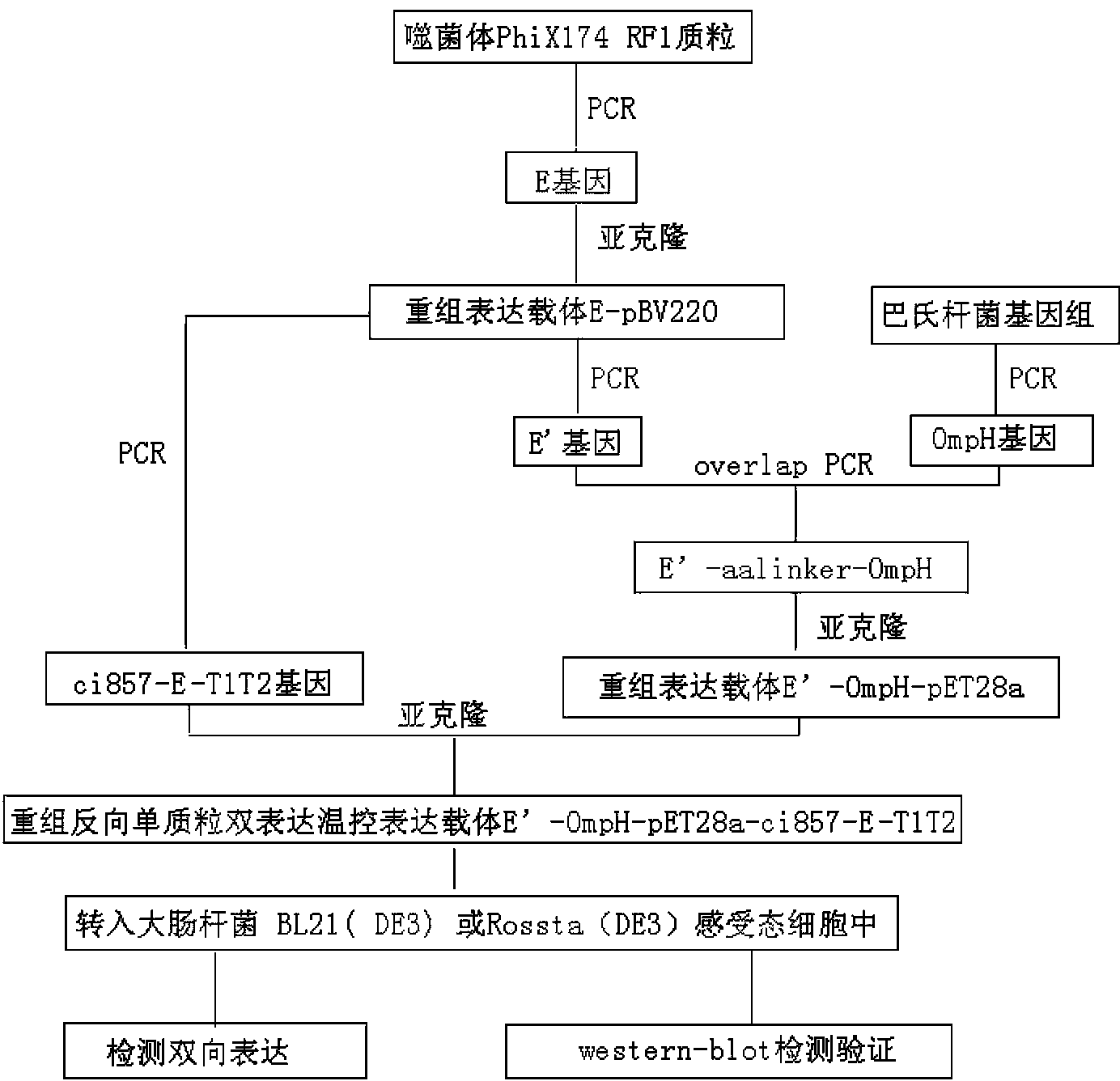
[0065] The construction of embodiment 2 recombinant plasmid E '-OmpH-pET28a
[0066] 1. Design and synthesis of primers
[0067] Since the two genes are connected in series using the overlap PCR method, two pairs of overlapping complementary primers are designed according to the constructed recombinant plasmids pBV220-E and OmpH, and a BamH I restriction enzyme site is introduced at the 5' end of the upstream primer F1 (underlined), and the 3' end of the downstream primer K2 was introduced with an XhoI restriction enzyme site (underlined). Using the recombinant prokaryotic expression plasmid pBV220-E as a template, primers F1 and F2 amplify the anchor gene E', using the genome of Pasteurella CVCC393 as a template, primers K1 and K2 amplify the OmpH gene, and then use primers F1 and K2 to amplify E'-OmpH fusion gene. The nucleotide sequences of the primers are as follows:
[0068] E'upstream primer F1: 5'-CGCGGATCCATGGTACGCTGGACTTTGTG-3'
[0069] E' downstream primer F2: 5'...
Embodiment 3
[0093] Example 3 Construction and expression of bidirectional expression vector E'-OmpH-pET28a-ci857-E-T1T2
[0094] 1. Primer design and synthesis
[0095] According to the published gene sequence of the expression system of pBV220, the application software Primer5.0 and Oligo6.0 were used to obtain the repressor protein ci857, E gene and terminator T of pBV220 1 T 2 Design a pair of primers C1 and T1 for the template to amplify the cleavage system ci857-E-T1T2 gene, introduce a SgrAI restriction endonuclease site (underlined part) at the 5' end of the upstream primer, and introduce a SphI restriction site at the 5' end of the downstream primer endonuclease site. Primers were synthesized by Shanghai Sangon Bioengineering Technology Service Co., Ltd.
[0096] The nucleotide sequences of the primers are as follows:
[0097] ci857 upstream primer C1: 5'-TACATACrCCGGyGTCAGCCAA ACGTCTCTTC-3'(SgrAI)
[0098] T1T2 downstream primer T1: 5'-ACATGCATGCGTTTGTAGAAACG CAAAAAGG-3'(Sph...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com