Traceless modification method of bacillus subtilis genome
A Bacillus subtilis, modification method technology, applied in microorganism-based methods, biochemical equipment and methods, bacteria and other directions, can solve the problems of high false positive rate, difficulty in screening positive recombinant strains, low absorption capacity, etc. The effect of chemical preparation process and excellent ability to transform exogenous DNA
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
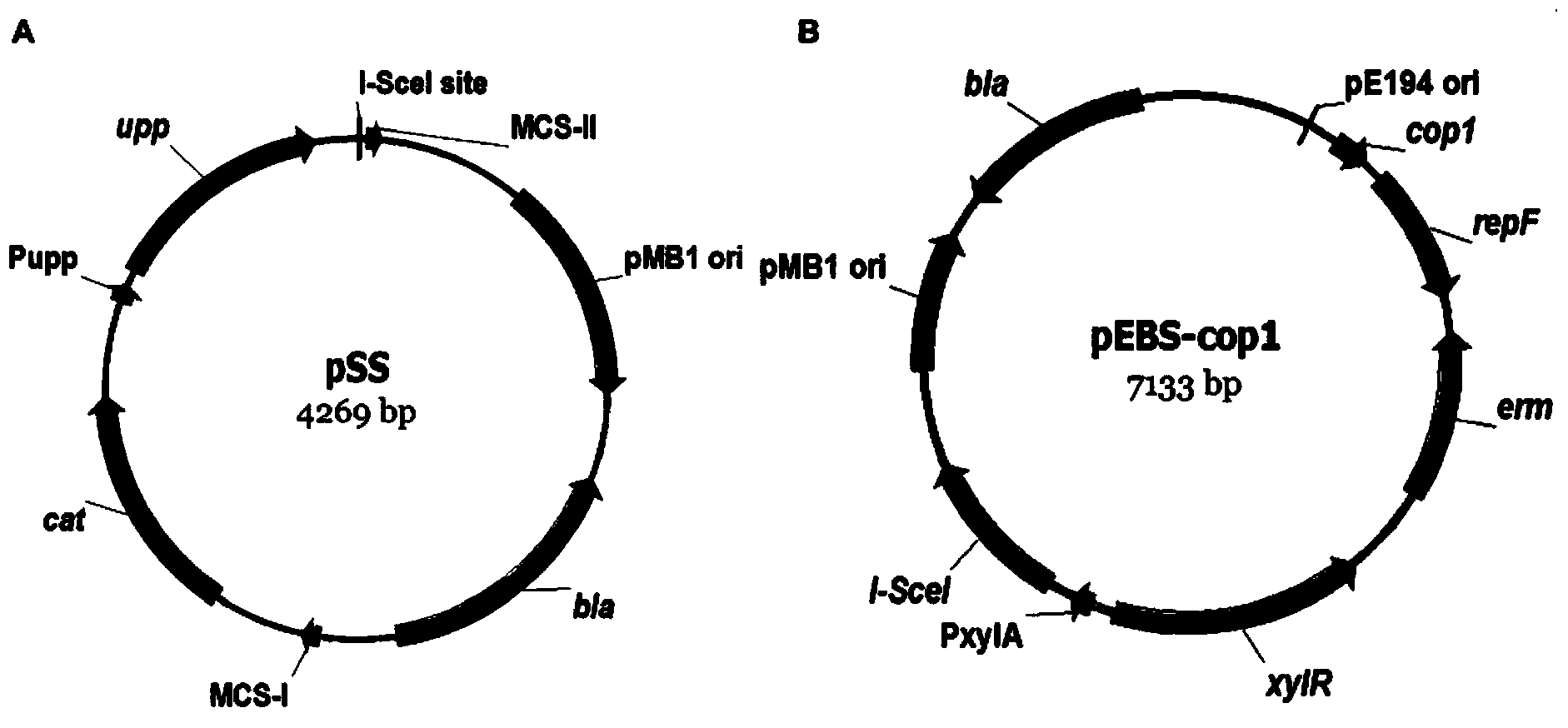
[0042] 1. Construction of the template plasmid pSS containing the I-SceI restriction site (see figure 1 A)
[0043] Use the PCR reaction to use the pC194 plasmid as a template to obtain the cat gene using the upstream and downstream primers pSS-P1 and pSS-P2, and use the B. subtilis168 genome as a template to obtain the upp gene using the upstream and downstream primers pSS-P3 and pSS-P4, and then use the above two A fragment was used as a template, and the recombinant fragment cat-upp was obtained by fusion PCR reaction using upstream and downstream primers pSS-P1 and pSS-P4. The recombinant fragment and the pUC18 (universal plasmid) plasmid were subjected to enzyme digestion, enzyme ligation, transformation, verification, etc., to obtain the basic operation plasmid pSS (see Table 2 for the primer sequences used).
[0044] 2. Construction of temperature-sensitive expression plasmid pEBS-copl (see map figure 1 b)
[0045] The erm gene was obtained by using the pE194 plasmid...
Embodiment 2
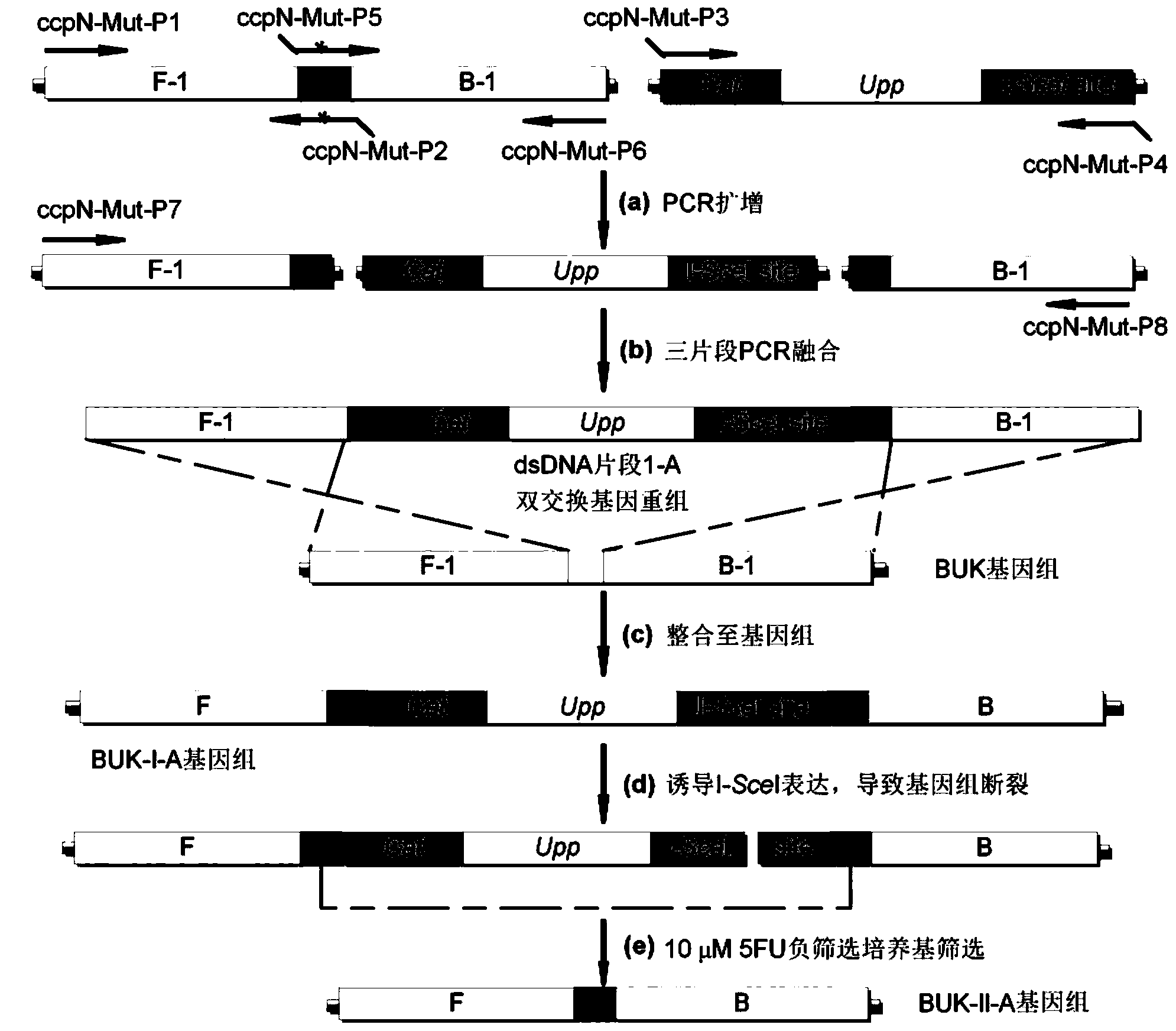
[0061] Example 2: Gene mutation introduced by traceless genetic manipulation (ccpN*G130T) see image 3
[0062](1) Using the plasmid pSS as a template, primers ccpN-Mut-P3 and ccpN-Mut-P4 as amplification primers, and PCR to obtain positive and negative screening boxes; using the B. subtilis168 genome as a template, using ccpN-Mut-P1 and containing The target gene mutation (G130T) ccpN-Mut-P2 was used as a primer, and the upstream homology arm F-1 (1379bp) containing the target mutation (G130T) DR sequence was obtained by PCR; B. subtilis168 was used as a template to contain the target gene mutation ( ccpN-Mut-P5 and ccpN-Mut-P6 of G130T) were used as primers, and the downstream homology arm B-1 (1339bp) containing the DR sequence of the target mutation (G130T) was obtained by PCR; the DR containing the target mutation (G130T) The upstream homology arm F-1 of the sequence, the positive and negative screening box and the downstream homology arm B-1 containing the DR sequence o...
Embodiment 3
[0068] Example 3: The introduction of gene knockout (ΔccpN) by traceless genetic manipulation see Figure 4
[0069] (1) Using the plasmid pSS as a template, primers ccpN-Del-P3 and ccpN-Del-P4 as amplification primers, PCR to obtain positive and negative screening boxes; using the B. subtilis168 genome as a template, using ccpN-Del-P1 and ccpN -Del-P2 was used as a primer, and the upstream homology arm F-2 (1110bp) containing DR sequence was obtained by PCR; B. subtilis168 genome was used as template, and ccpN-Del-P5 and ccpN-Del-P6 containing DR sequence were used as Primers, PCR to obtain the downstream homology arm B-2 (1140bp) containing the DR sequence; the upstream homology arm F-2 containing the DR sequence, the positive and negative screening box and the downstream homology arm B- 2. Using ccpN-Del-P7 and ccpN-Del-P8 as primers, obtain the target dsDNA fragment 1-B by fusion PCR;
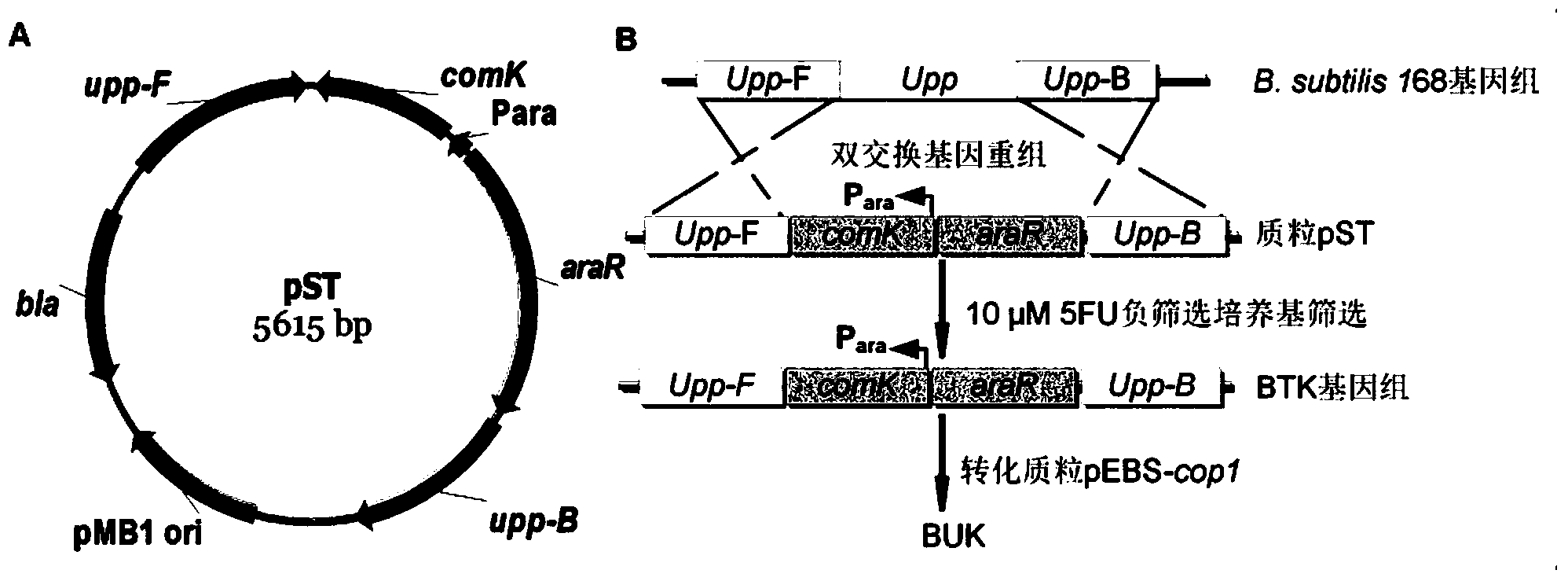
[0070] (2) Inoculate BUK in 5mL LB liquid medium, culture overnight at 37°C and 220rp...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com