RBD (receptor binding domain) segment in MERS-CoV (Middle East respiratory syndrome coronavirus) membrane protein and coding gene and application thereof
A protein and residue technology, applied to the RBD fragment in the MERS-CoV virus membrane protein and its encoding gene and application field, can solve the problem of undetermined source and host, and achieve the effect of extensive theoretical guidance value
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0045] Embodiment 1, the preparation of RBD fragment
[0046] 1. Construction of recombinant plasmids
[0047] FastBac TM The dual vector itself does not have a signal peptide secreted extracellularly, and the RBD fragment itself is a secreted protein, so the coding sequence of the signal peptide needs to be connected to the vector in advance, and the signal peptide will be excised during the process of protein expression and secretion in insect cells.
[0048] 1. Synthesize the double-stranded DNA molecule shown in sequence 4 of the sequence listing (the double-stranded DNA molecule shown in sequence 4 of the sequence listing encodes the gp67 signal peptide).
[0049] 2. Using the double-stranded DNA molecule synthesized in step 1 as a template, perform PCR amplification with a primer pair composed of F1 and R1 to obtain a PCR amplification product.
[0050] F1: 5'-cgg AGATCT ATGCTACTAGTAAATCAG-3';
[0051] R1: 5'-GCG GAATTC GC CGCAAAGGCAGAATGCGC-3'.
[0052] 3. Dige...
Embodiment 2
[0091] Example 2, Preparation of DPP4 Extracellular Region Fragments
[0092] 1. Construction of recombinant plasmids
[0093] 1. Synthesize the double-stranded DNA molecule shown in sequence 7 of the sequence listing.
[0094] 2. Using the double-stranded DNA molecule synthesized in step 7 as a template, perform PCR amplification with a primer pair composed of F3 and R3-1 to obtain a PCR amplification product.
[0095] F3: 5'-cgc ggatcc cAGTCGCAAAACTTACACTCT-3';
[0096] R3-1: 5'-ATACGC GTC GAC CTAatggtggtgatggtggtg AGGTAAAGAGAAACATTGTTTTATG-3'.
[0097] 3. Using the double-stranded DNA molecule synthesized in step 1 as a template, perform PCR amplification with a primer pair composed of F3 and R3-2 to obtain a PCR amplification product.
[0098] R3-2: 5'-ATACGC GTC GAC CTA AGGTAAAGAGAAACATTGTTTTATG-3’
[0099]4. Digest the PCR amplified product in step 2 with restriction endonucleases BamHI and SalI, and recover the digested product.
[0100] 5. Digest the recombi...
Embodiment 3
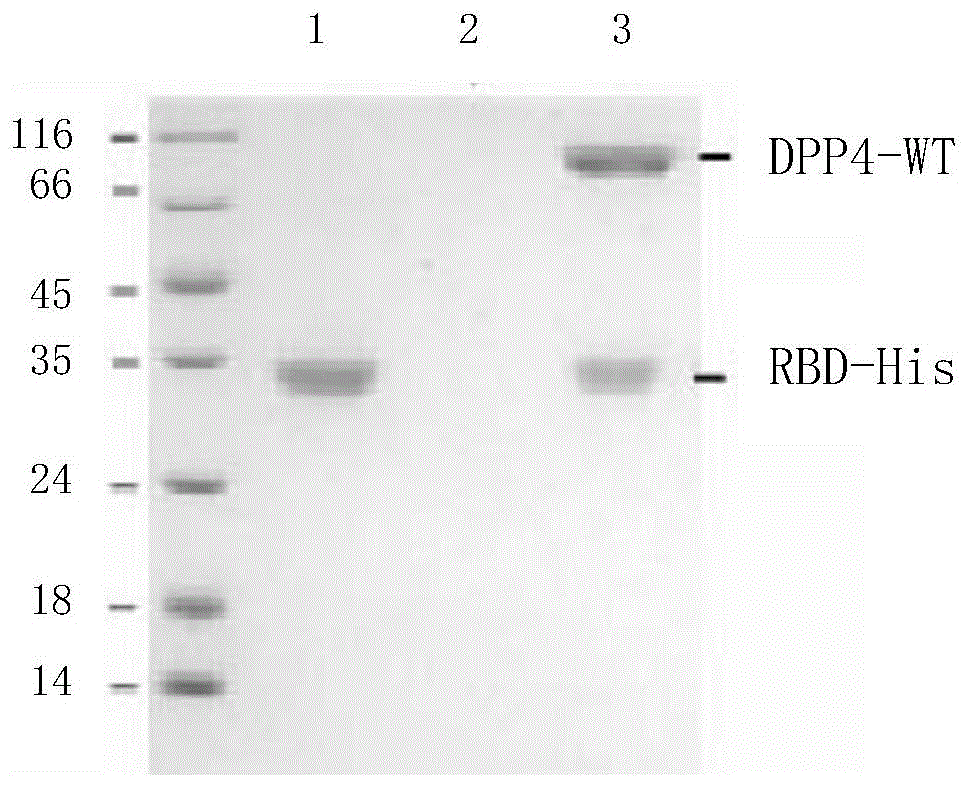
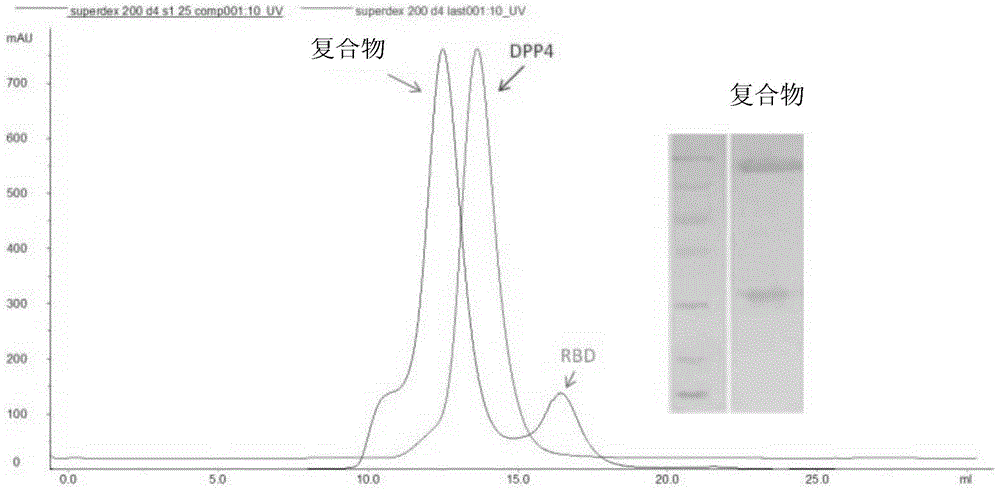
[0119] Example 3, the binding experiment of RBD and DPP4
[0120] 1. Co-expression and pull-down experiments
[0121] 1. The DPP4-WT-P1 generation virus solution prepared in Example 2 and the RBD-His-P1 generation virus solution prepared in Example 1 were respectively infected or co-infected at a density of 2×10 6 cells / mL of Sf9 cell fluid (when infecting separately, the volume ratio of the virus fluid to the cell fluid is 1:100; when co-infecting, the two virus fluids are mixed in equal volumes, and the volume ratio of the mixed virus fluid to the cell fluid is 1:100 ), cultured at 27°C, 110rpm for 72h, then centrifuged at 4000rpm for 5min, and the supernatant was taken.
[0122] 2. Add nickel-coated affinity chromatography column material to the supernatant obtained in step 1, shake at 4°C and 30rpm for 3h, then centrifuge at 4000rpm for 5min, and take the precipitate.
[0123] 3. Add Buffer1 containing 20mM imidazole to the precipitate obtained in step 2 for washing, the...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com