Multipurpose DNA segment enrichment method used for next generation sequencing
A technology of sequencing and fragmentation, which is applied in the field of genetic engineering, can solve problems such as indistinguishability, large amount of initial genomic DNA, and interference of results, and achieve simple and clear data processing, saving time and economic costs, and lowering the requirements for sequencing depth Effect
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0044] Example 1. Preparation and sequencing of sequencing templates for Illumina GA sequencing
[0045] 1. Preparation method of sequencing template for Illumina GA sequencing
[0046] In this embodiment, the method for preparing a sequencing template for Illumina GA sequencing specifically includes the following steps:
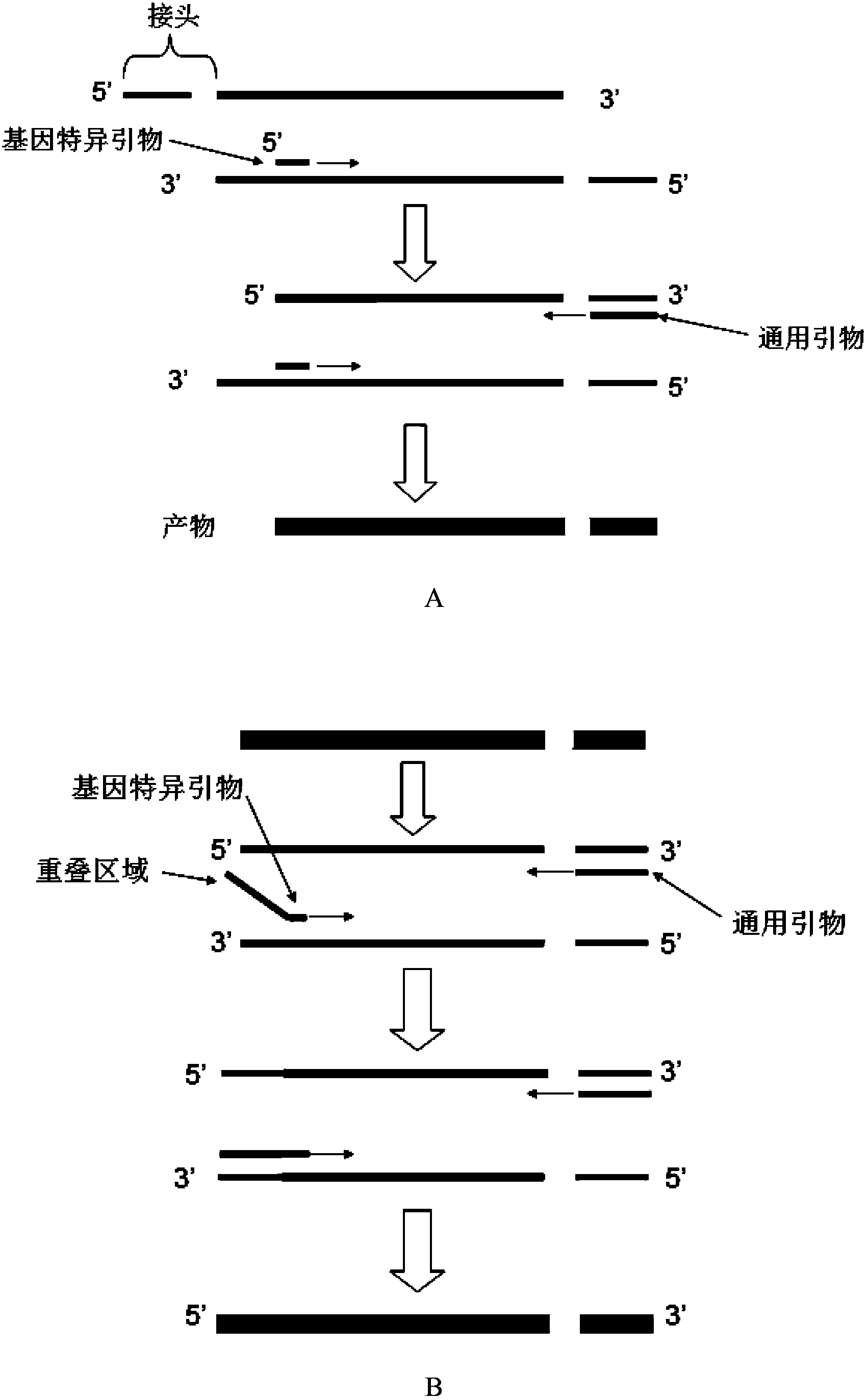
[0047] (1) The DNA sample with the site to be sequenced is fragmented, end repaired and filled in sequence, A is added to the 3' end, and the adapter is connected to obtain a template for the first PCR reaction;
[0048] The upstream nucleotide sequence of the site to be sequenced is known, and there is a specific region corresponding to the PCR specific primer;
[0049] The linker is a double-stranded DNA consisting of a long chain and a short chain with a blunt end and a 5' sticky end at one end, and the 5' end of the long chain protrudes;
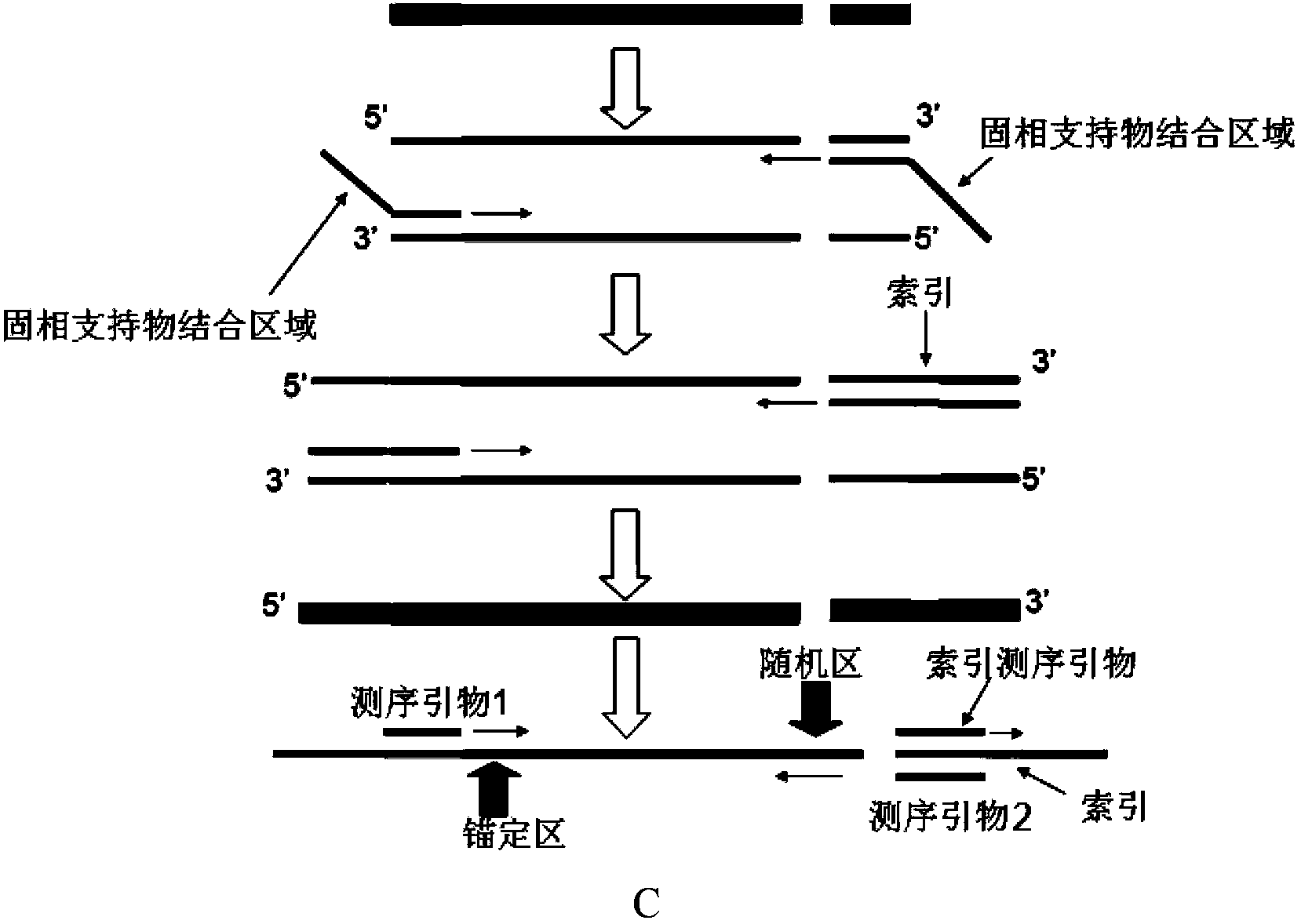
[0050] (2) Using specific primer 1ros and universal primer 1 to perform the first PCR amplification on the templat...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com