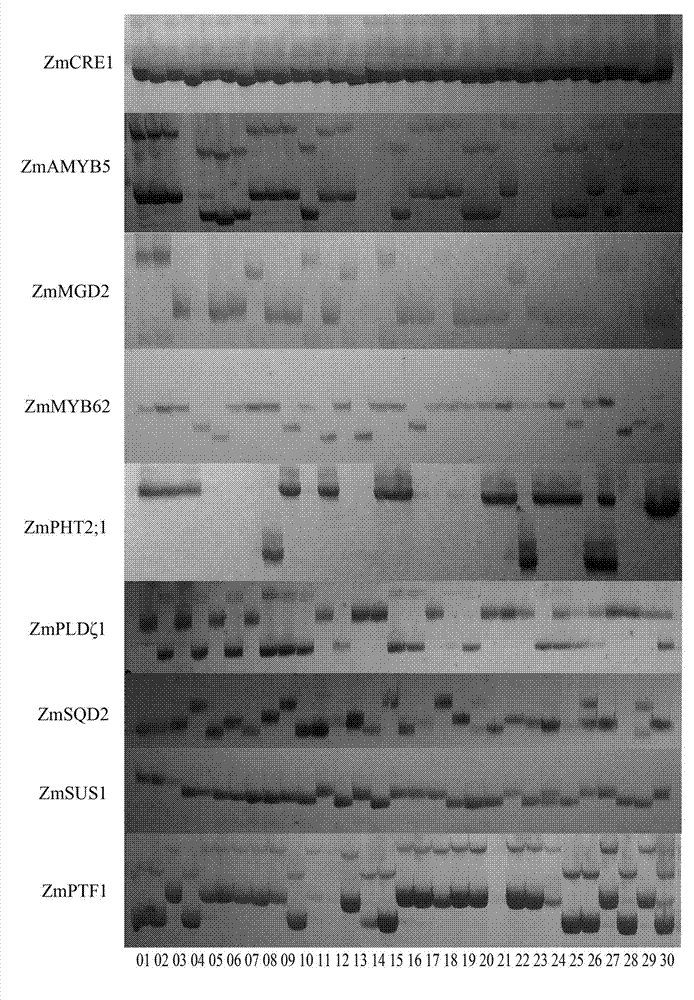
Maize phosphate starvation responses intron length polymorphism marker for corn
A length polymorphism and intron technology, applied in the field of molecular biology and genetic breeding, can solve the problem of high cost, and achieve the effect of low cost, strong applicability and simple operation
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0028] Example 1 ZmCRE1 Gene Intron Length Polymorphism Marker Development
[0029] 1. Genomic DNA extraction
[0030] Genomic DNA was extracted from 30 seedling leaves of maize inbred lines (Table 1, provided by the Maize Research Institute of Sichuan Agricultural University) using the CTAB method.
[0031] serial number Inbred pedigree serial number Inbred pedigree 01 178 American hybrid 78599 selection line 16 Qi205 (VeiAi141×ZhongXi017)×Population70 02 9782 american hybrid 17 Dan599 American hybrid 78599 selection line 03 B73 BSSS group selection system 18 Chang7-2 Huang Zaosi×Wei 95×S901 04 MO17 CL187-2×C103 19 Dan340 Bone Brigade 9 × Pale Corn (Radiation) 05 CML166 P66C1F215-4-1-2-BB-2-BBB 20 S37 SUWAN1 group 06 (CML-306)-B SINT.AM.TSR-19-1-2-3-1-BB-f 21 Jiao51 Guizhou Local Breeding Lines 07 (CML-473)-B P31C4S5B-23-#-#-4-BBBB 22 Dan598 American hybrid 78599 selectio...
Embodiment 2
[0045] Example 2 ZmAMYB5 gene intron length polymorphic marker development
[0046] 1. Extract genomic DNA, same as Example 1.
[0047] 2. PSR-ILP specific primer design
[0048] The gene sequence and cDNA sequence of the ZmAMYB5 gene (GRMZM2G058310) were downloaded in the MaizeGDB database. After Blast sequence alignment and sequence analysis, specific primers were designed for the second intron of ZmAMYB5 gene. The upstream and downstream primers were designed respectively in the 2nd exon region and the 3rd exon region spanning the 2nd intron, and after specific primer screening and optimization, the upstream primer ZmAMYB5-ILP-F and downstream primer ZmAMYB5-ILP-R were obtained ,details as follows:
[0049]ZmAMYB5-ILP-F: 5’ TCTTCTACACCAACCGGAGTG 3’
[0050] ZmAMYB5-ILP-R: 5’ AGTCCCACCTCAATGTCCAC 3’
[0051] 3. PCR amplification and detection
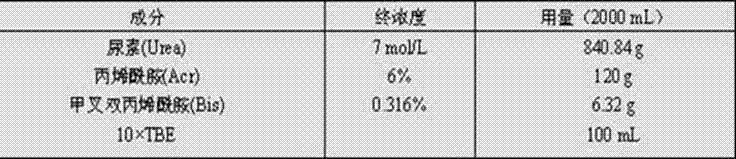
[0052] 3.1 PCR reaction system Same as Example 1.
[0053] 3.2 PCR reaction procedure
[0054] Pre-denaturation at 94°C for ...
Embodiment 3
[0056] Example 3 ZmMGD2 Gene Intron Length Polymorphism Marker Development
[0057] 1. Extract genomic DNA, same as Example 1.
[0058] 2. PSR-ILP specific primer design
[0059] The gene sequence and cDNA sequence of the ZmMGD2 gene (GRMZM2G178892) were downloaded in the MaizeGDB database. After Blast sequence alignment and sequence analysis, specific primers were designed for the 8th intron of ZmMGD2 gene. The upstream and downstream primers were respectively designed in the 8th exon region and the 9th exon region spanning the 8th intron, and after specific primer screening and optimization, the upstream primer ZmMGD2-ILP-F and downstream primer ZmMGD2-ILP-R were obtained ,details as follows:
[0060] ZmMGD2-ILP-F: 5’ CTGGACCAGGTACCATTGCT 3’
[0061] ZmMGD2-ILP-R: 5’ CGGGCAACAAGACTAGCAG 3’
[0062] 3. PCR amplification and detection
[0063] 3.1 PCR reaction system, same as Example 1.
[0064] 3.2 PCR reaction procedure
[0065] Pre-denaturation at 94°C, 5 min; denat...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com