High-catalytic-activity mutant enzyme for D-allulose 3-epimerase and application thereof
An epimerase, high catalytic activity technology, applied in the field of genetic engineering of enzymes, can solve the problems of difficult to obtain, low content and so on
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0022] Example 1: DPE enzyme site-directed mutation analysis and mutant preparation method
[0023] Through the analysis of the 3D spatial structure of DPE enzyme, it was found that Y68 is located in the active pocket. By changing the amino acid at position 68, the shape of the hydrophobic pocket can be changed, thereby changing the enzyme activity. The study found that replacing the 68-position tyrosine Tyr with isoleucine Ile can increase its catalytic activity.
[0024] Construction of the pet22b(+)-Y68I mutant plasmid by rapid PCR site-directed mutagenesis;
[0025] The construction of the pet22b(+)-Y68I mutant plasmid uses the pet22b(+)-cb-dpe plasmid as a template, and uses the Y68I mutant primer to obtain a product with a size of about 6.4kbp by PCR once. Dpn After I treatment, Escherichia coli DH5α competent cells were transformed, and transformants were selected for sequencing verification.
[0026] Sequencing verification results showed that there was no random m...
Embodiment 2
[0042] Embodiment 2: Clostridia ( Clostridium boltae ) A method for expressing and purifying mutant enzymes of DPE.
[0043] The mutant plasmid pet22b(+)-Y68I verified by sequencing was transformed into E. coli BL21(DE3) cells, and the positive transformants were picked and cultured in LB medium at 37°C, 200rpm, shaking overnight, and then cultured in LB medium at 37°C After 3-4 hours until the OD value is 0.6-0.8, cool down to 25°C and add IPTG at a final concentration of 0.8mM to induce for 8 hours.
[0044] The fermentation broth was centrifuged at 10,000 rpm for 20 min at 4°C, and the cells were collected. Add 15 mL Binding Buffer (50 mM Na 2 HPO 4 , 50 mM NaH 2 PO 4 , 500 mM NaCl, adjust the pH to 7.4) to fully resuspend the bacteria, then place the centrifuge tube in an ice bath and put it into an ultrasonic cell disruptor. The conditions for ultrasonic disruption are: working time 1 s, stop time 2 s, total 20 min. Centrifuge the broken solution at low temperature...
Embodiment 3
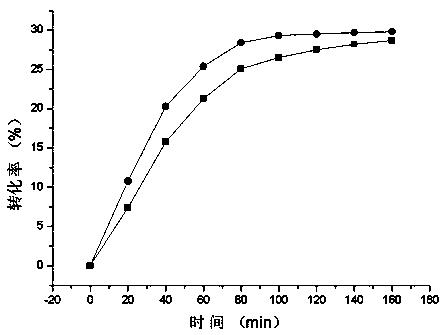
[0046] Example 3: Determination of reaction kinetic constants of DPE enzyme at 55°C
[0047] 1. DPE enzyme activity assay method: Add 700 μL of 100 g / L D-fructose to 1 mL of reaction system, 200 μL of diluted enzyme solution, and 100 μL of Co at a final concentration of 1 mM 2+ ion. Incubate at 55°C for 5 min, then boil for 10 min to terminate the enzyme reaction.
[0048] 2. Study on kinetic constants of enzyme reaction
[0049] D-fructose (10-200 mM) solutions with different substrate concentrations were prepared, and the enzyme activity was measured at 55°C and pH 7.0, and the regression line equation was obtained by the Lineweaver-Burk double reciprocal method, where the line and X The intersection of the axes is -( Km ) -1 , the intersection point with the Y axis is (Vm) -1 , while measuring the concentration of the purified enzyme protein, in order to obtain k cat value.
[0050] Table 1. Kinetic constants of enzymatic reactions K m and catalytic constant k c...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com