Method for cloning unknown flanking sequences of PS1 gene 5' of dianthus caryophyllus
A technology of unknown sequence and cloning method, applied in the field of bioengineering, can solve the problem of low expression of carnation PS1 gene, and achieve the effect of saving manpower, low cost and strong pertinence
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
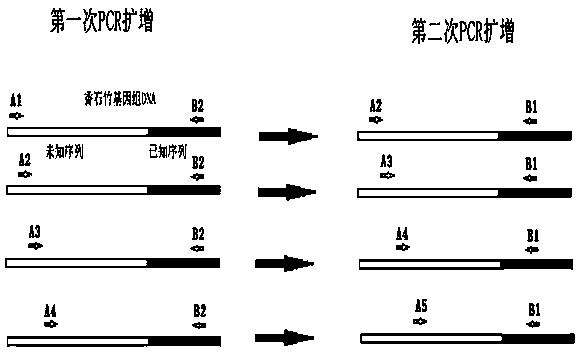
[0067] The design of embodiment 1 degenerate primer and specific primer
[0068] (1) Design of degenerate primers for amplifying the unknown sequence on the 5' flank of PS1 gene: through homologous comparison of PS1 gene, design 5 degenerate primers according to the conserved sequence, named A1, A2, A3, A4 and A5 , the base sequence of the A1 primer is shown in SEQ ID NO:1. The base sequence of the A2 primer is shown in SEQ ID NO:2. The base sequence of the A3 primer is shown in SEQ ID NO:3. The base sequence of the A4 primer is shown in SEQ ID NO:4. The base sequence of the A5 primer is shown in SEQ ID NO:5.
[0069] (2) Based on the middle fragment of the cloned PS1 gene of Carnation, two specific primers were designed, named B1 and B2 respectively. The base sequence of the B1 primer is shown in SEQ ID NO:6. The base sequence of the B2 primer is shown in SEQ ID NO:7.
[0070] The above primers were synthesized by Shanghai Sangon Bioengineering Technology Co., Ltd.
Embodiment 2
[0071] Example 2 Cloning of Carnation PS1 Gene 5' flanking Unknown Sequence
[0072] (1) Carnation genomic DNA was extracted by CTAB method, amplified by nested PCR, using the extracted Carnation genomic DNA as a template, and A1 and B2 as primers for the first round of PCR amplification: the total reaction volume was 25 μL, of which 10×Ex TaqBuffer2.5μL, 25mM MgCl 2 1.5 μL, 2.5 mM dNTP 2 μL, 10 μM A1 primer 1 μL, 10 μM B2 primer 1 μL, 5 U / μL Ex Taq 0.15 μL, template DNA 1 μL, the rest is sterile water;
[0073] The amplification procedure is:
[0074] (1), 1min at 95°C,
[0075] (2), 94°C for 30s,
[0076] (3), 60°C for 1min,
[0077] (4), 72°C 2min30s,
[0078] (5), from step (2) to step (4), carry out a total of 10 rings,
[0079] (6), 94°C for 30s,
[0080] (7) 2min at 25°C, rising to 72°C at 0.5°C / s,
[0081] (8), 72°C 2min30s,
[0082] (9), 94°C for 30s,
[0083] (10), 58°C for 1min,
[0084] (11), 72°C for 2min30s,
[0085] (12), from steps (9) to (11) a tot...
Embodiment 3— Embodiment 5
[0108] Example 3-Example 5 are all clones of the unknown sequence of the 5' flank of Carnation PS1 gene, except for the operations listed in Table 1, the rest of the operations are the same as in Example 2, and will not be repeated. See Table 1 for details.
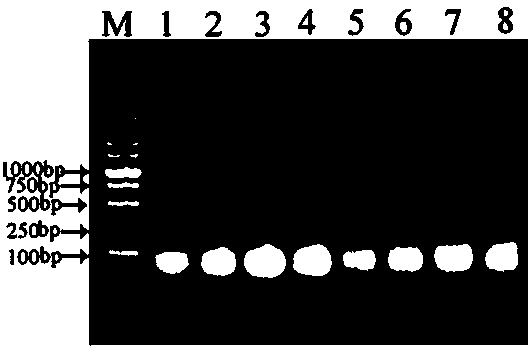
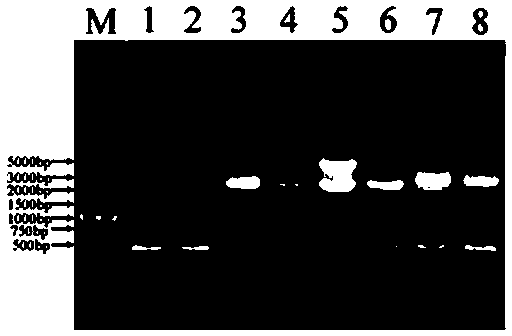
[0109] Embodiment 1-Example 5 adopts the nested PCR amplified agarose gel electrophoresis to detect PCR product fragment results see Figure 2-Figure 3 ,Table 2.
[0110] Table 2 embodiment 1-embodiment 5 adopts nested PCR amplified agarose gel electrophoresis to detect PCR product fragment result
[0111]
[0112] The time used in each of the above embodiments is 6-8 hours, and the sequencing results show that the obtained fragment is the target fragment including the known sequence, and the method of the present invention can be conveniently, quickly, efficiently and accurately cloned into Carnation PS1 gene 5 'flanking unknown sequences.
[0113] Table 1 Example 3-Example 5 Carnation PS1 gene 5' flanking unknown ...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com