Specific sub sequence for separating Ac/Ds flanking sequence and separation method thereof
A technology of linker sequence and flanking sequence, applied in the field of specific linker sequence for separating Ac/Ds flanking sequence and separation thereof
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0028] 1. Acquisition of linkers: According to the sequence of the blunt-end linkers of Clontech's genomewalking kit, the present invention designs linker sequences suitable for producing 5' protruding ends, wherein linker 1 is quoted from the instructions of Clontech's genomewalking kit, which is suitable for different Linker 2 of the restriction endonuclease was designed by us based on the sequence of linker 1.
[0029]
[0030] Dissolve the adapter 1 and adapter 2 synthesized by the company with sterile water to a final concentration of 200 μM, and prepare the adapter solution in a 0.5ml centrifuge tube according to the following formula:
[0031] Adapter 1 (200μM) 10μl
[0032]Adapter 2 (200μM) 10μl
[0033] 60mM Tris-HCl pH7.5, 150mM NaCl 10μl
[0034] The prepared solution was incubated in a 94°C water bath for 2 minutes, and then naturally cooled to room temperature in a water bath to anneal the two single-stranded DNAs to form a partially double-stranded linker. ...
Embodiment 2
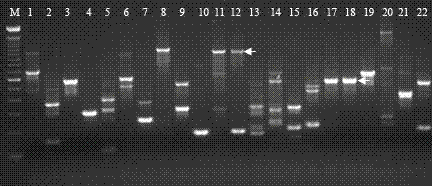
[0045] Example 2: Separation of specific bands co-segregated with mutant phenotypes
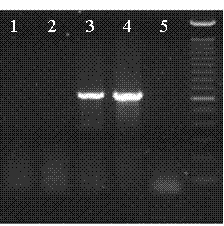
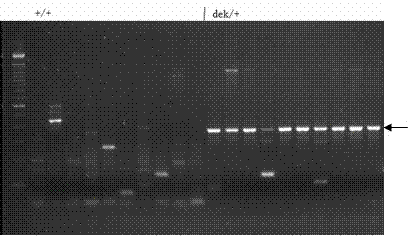
[0046] In the isolated population of the corn kernel mutant shu00067, wild-type homozygous plants (+ / +) and wild-type heterozygous plants (dek / +) were selected to extract DNA, and the genomic DNA was digested with SalI, using the method used in Example 2 Ac / Ds flanking sequences were isolated and a band was identified that co-segregated with the dek mutant phenotype ( image 3 ). Primers were designed by sequencing the flanking sequences of this PCR band, and the bands were only amplified in mutant grains (- / -) and wild-type heterozygous (+ / -) with Ac-terminal-specific primers and flanking sequence-specific primers , while no corresponding product was amplified in wild-type control W22 and wild-type homozygous plants ( Figure 4 ), the results indicated that the assigned flanking sequences were genuine and co-segregated with the dek mutant phenotype.
[0047] Shanghai University
[0048]...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com