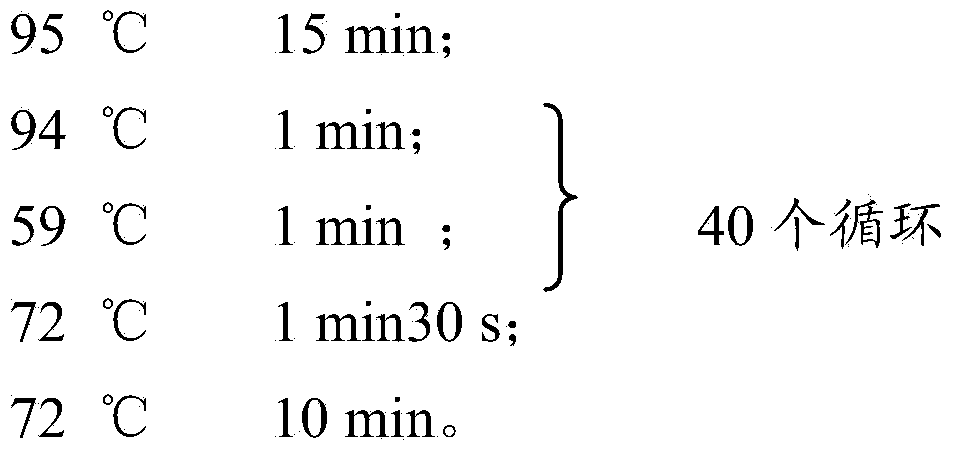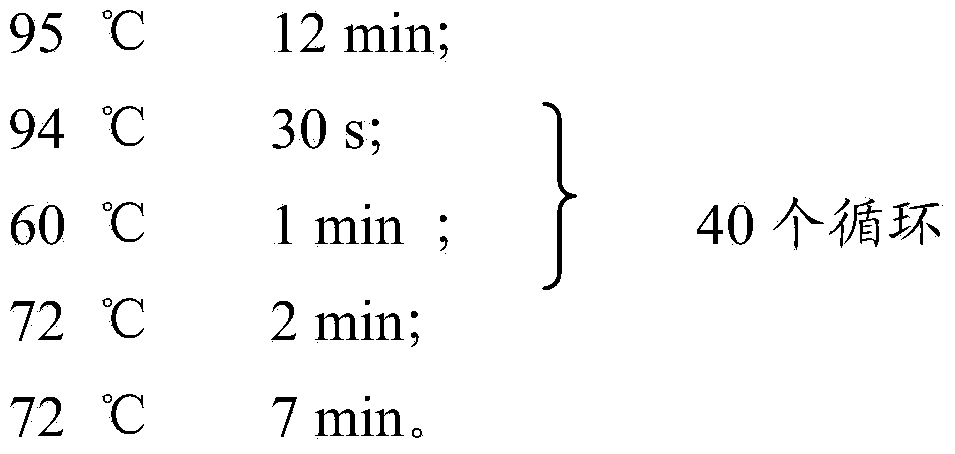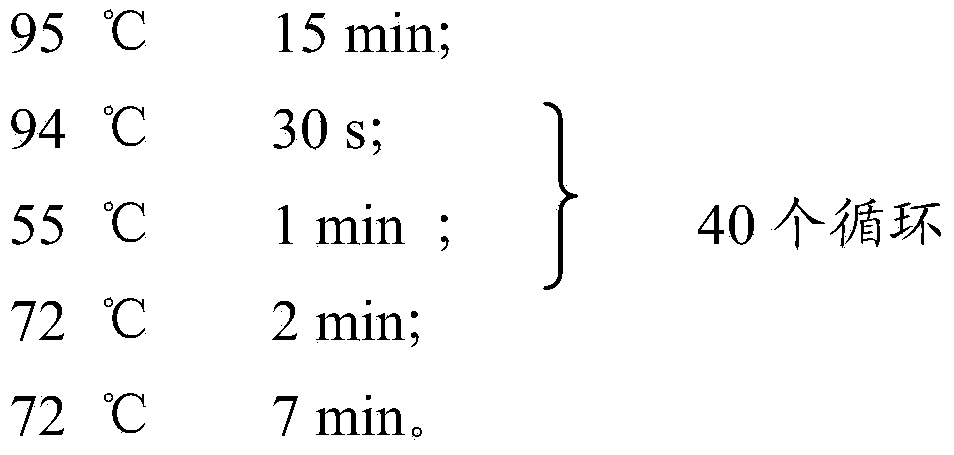Genotype detection primer group and kit for mycobacterium tuberculosis
A technology for Mycobacterium tuberculosis and genotyping, applied in microorganism-based methods, microorganisms, recombinant DNA technology, etc., and can solve problems such as differences in discrimination ability, low typing index, poor stability and repeatability
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0086] Example 1: Extraction of Mycobacterium tuberculosis Genomic DNA
[0087] Take 48 strains of Mycobacterium tuberculosis to be tested, pick 2-3 colonies for each strain, put them into centrifuge tubes respectively, add 200 μL TE buffer solution and add SDS to make the final concentration 1%, and store at 85°C-90°C After cooking at ℃ for 10 minutes, the genomic DNA of Mycobacterium tuberculosis was extracted according to the bacterial DNA extraction kit of Takara Company. The obtained genomic DNA was stored at -20°C before testing.
Embodiment 2
[0088] Example 2: PCR amplification of different VNTR sites of the strains to be tested
[0089] With 22 pairs of primers in the Mycobacterium tuberculosis genotyping detection primer set provided by the present invention, PCR amplification is carried out to the VNTR sites in 48 strains to be tested respectively:
[0090] The amplification system is:
[0091]
[0092] The PCR reaction conditions for the sites MIRU4, MIRU10, MIRU31, MIRU39, MIRU26, MIRU2, MIRU24, MIRU16, MIRU40, MIRU20, MIRU23 and MIRU27 are:
[0093]
[0094] The PCR reaction conditions for sites ETR A, QUB-18, ETR B, ETR C, QUB-11a and QUB-26 are:
[0095]
[0096] The PCR reaction conditions for sites QUB-3232, QUB-3336, QUB-1895, QUB-4156 and QUB-2163 are:
[0097]
[0098] The PCR reaction conditions for sites Mtub34, Mtub29, Mtub39, Mtub30, Mtub21 and Mtub4 are:
[0099]
[0100] After PCR amplification, the PCR products of 22 VNTR sites in 48 strains were obtained.
Embodiment 3
[0101] Embodiment 3: Electrophoresis and fragment copy number identification of PCR products
[0102] The PCR products of each VNTR site in the 48 strains were detected by capillary electrophoresis to determine the fragment size of the PCR products.
[0103] The conditions of capillary electrophoresis are as follows:
[0104] The sample volume is 5 μL, voltage: 100V-240V, 50 / 60Hz; working temperature: 10°C-30°C; working humidity: 10%-75%. The cartridges used are QX DNA Screening Cartridge and QX DNA High Resolution Cartridge respectively.
[0105] Running program: Use the program that comes with the system software, the detection time is 450s, and the sampling time is 10s.
[0106] According to the length of the product, the copy number of the VNTR locus is converted. If the copy number of each VNTR locus is consistent, it is judged as the same genotype;
[0107] Wherein, the copy number of the VNTR site=PCR product size / length of each copy of the VNTR site. The calculatio...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com