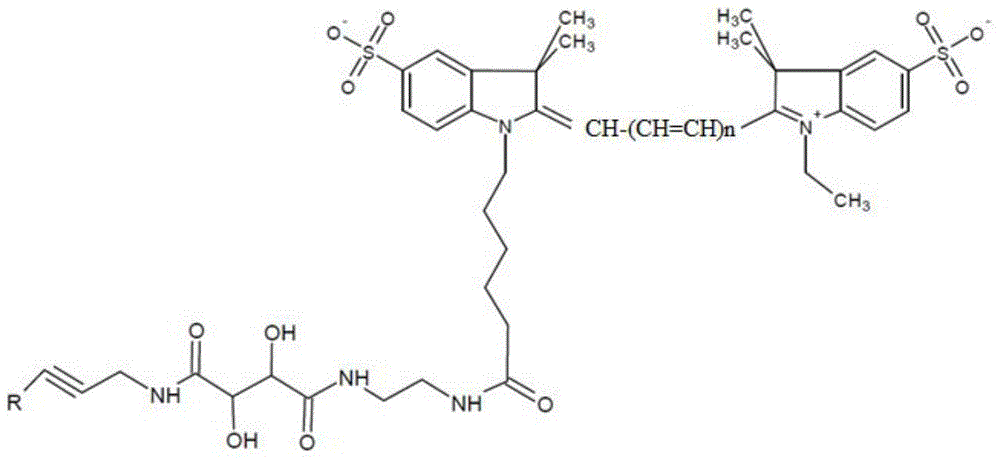
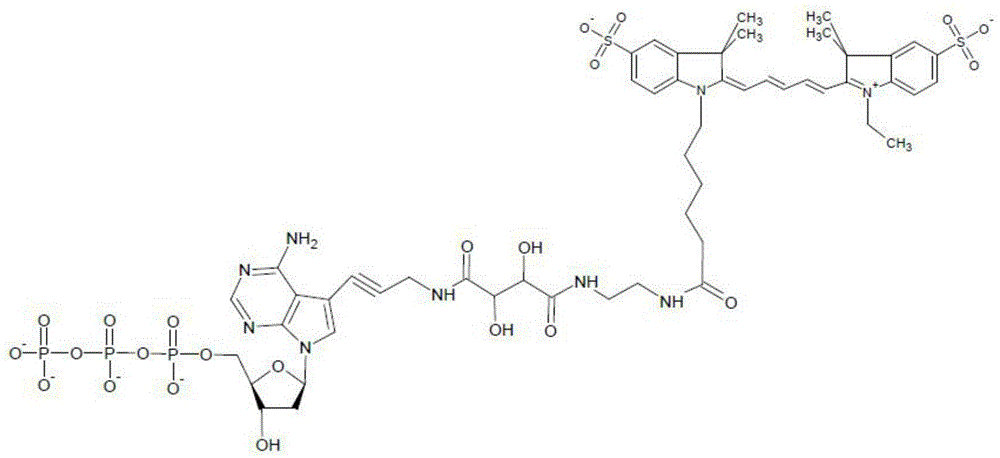
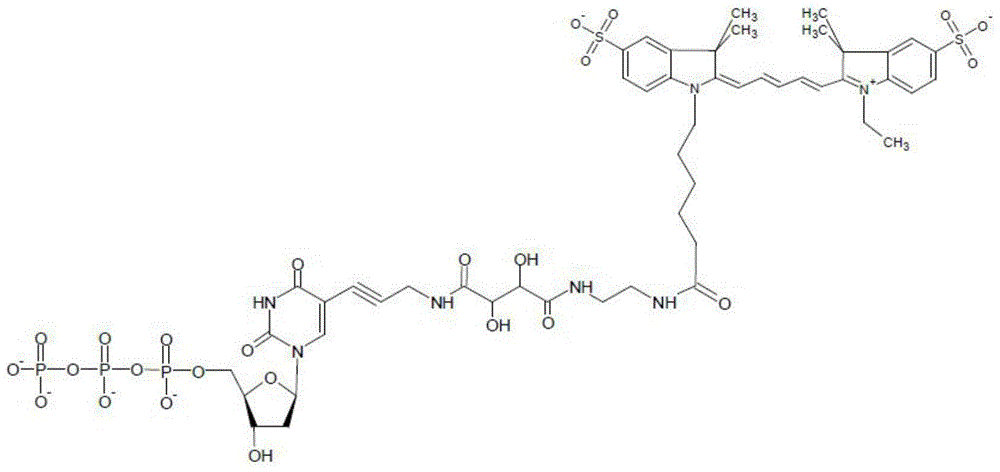
A special modified nucleotide and its application in high-throughput sequencing
A nucleotide and high-throughput technology, applied in the biological field, can solve problems such as limited read length, large uncertainty, and limited applications
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0063] Example 1: Sequencing of specially modified nucleotide codes in the Yanhuang No. 1 genome
[0064] 1. Whole-genome template preparation: use ultrasonic method to break the target genome into base fragments of 100-500bp, and add a pair of universal linkers P1 and P2 whose sequence is known to both ends of the DNA fragment under the action of ligase; Then the 180-220bp DNA fragment was cut and purified by gel electrophoresis, and pre-amplified for 8 cycles; then these pre-amplified DNA fragments were subjected to emulsion PCR reaction with magnetic beads immobilized with a complementary sequence of one of the linkers, A large number of monoclonal magnetic beads with long target genomic DNA fragments are obtained, and the genome sequencing templates are obtained by denaturation, and finally these magnetic beads amplified with fragmented DNA templates are immobilized on a microfluidic chip.
[0065] 2. Sequencing primer hybridization: hybridize the template strand on the mo...
Embodiment 2
[0080] Example 2: Specific modified nucleotide-encoded sequencing of the Saccharomyces cerevisiae genome
[0081] 1. Whole-genome template preparation: Saccharomyces cerevisiae genome is broken into base fragments of 100-500 bp by ultrasonic method, and a pair of universal linkers P1 and P2 with known sequences are added to both ends of the DNA fragment under the action of ligase ; Then the 180-220bp DNA fragment was cut and purified by gel electrophoresis, and pre-amplified for 8 cycles; then these pre-amplified DNA fragments were subjected to emulsion PCR reaction with magnetic beads immobilized with a complementary sequence of one of the linkers , obtain a large number of monoclonal magnetic beads with long target genomic DNA fragments, and denature them to obtain genome sequencing templates, and finally immobilize these magnetic beads amplified with fragmented DNA templates on a microfluidic chip.
[0082] 2. Sequencing primer hybridization: hybridize the template strand o...
PUM
| Property | Measurement | Unit |
|---|---|---|
| fluorescence | aaaaa | aaaaa |
Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com