Bleomycin resistance reporter gene mutant, and preparation method and application thereof
A bleomycin and reporter gene technology, applied in the field of molecular biology, can solve the problems of whole gene range detection, low efficiency, inability to locate off-target effects, etc.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0040] Example 1 Construction of Bleomycin Resistance Reporter Gene Mutants with PPICZαA and pEGFP-N3 Plasmids
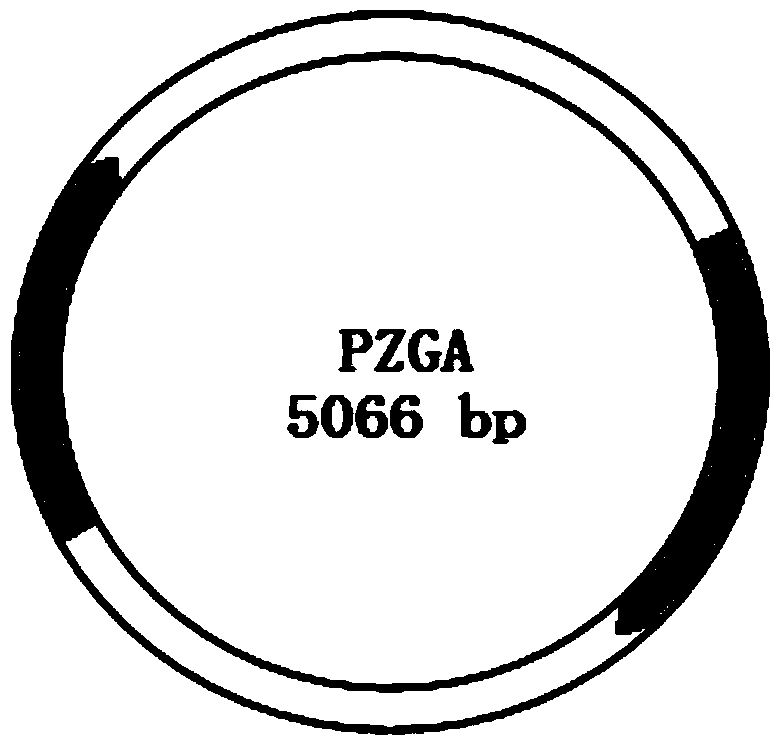
[0041] (1). Prepare the reporter plasmid pZGA that can be used to explore the concentration of bleomycin used in specific experiments.
[0042]In order to achieve the above object, the technical scheme of the present invention is: insert the bleomycin resistance gene coding sequence before the GFP reading frame of the pEGFP-N3 plasmid, and remove its stop codon at the same time to fuse the bleomycin resistance gene reading frame and GFP reading frames. Specifically include the following steps:
[0043] 1. Using the PPICZαA plasmid as a template,
[0044] Primer 1: CGATCCGCTAGCATGGCCAAGTTGACCAGTGCCGTTC (SeqNo.1)
[0045] Primer 2: TGGCTGGATCCGTCCTGCTCCTCGGCCACGAAG (Seq No.2)
[0046] A PCR amplification kit (Thermoscientific Company) was used to amplify the sequence Seq No. 16 corresponding to the target fragment of the bleomycin resistance gene using the above p...
Embodiment 2
[0070] Embodiment 2 constructs bleomycin resistance reporter gene mutant with PMD-19 and pPICαA plasmid
[0071] (1) Construction of bleomycin and ampicillin double resistance plasmid pZAA
[0072] 1. Design and synthesize specific primers for overlap extension PCR to obtain the bleomycin resistance gene 1.1 Using the PMD-19 plasmid as a template,
[0073] Primer 4: 5'-CCTCTGACTTGAGCGTCGATTTTTGTG-3'; (Seq No.4)
[0074] Primer 5: 5'-GTCAACTTGGCCATAGCTGTTTCCTGTGTGAAATTGTTATCCG-3'(Seq No.5)
[0075] Perform PCR amplification to obtain the target product fragment 1; wherein, each 25 μL PCR reaction system includes: 2.5 μL of 10×Pfu Buffer with MgSO4, 0.3 μL of Pfu Polymerase (2.5U / μL), 0.5 μL of dNTP Mix (10mM each ), the two specific primers were 5μM and 5μM respectively, the template was 40ng, and the rest was water. The PCR reaction conditions were: pre-denaturation at 95°C for 5min; denaturation at 95°C for 20S, annealing at 58°C for 20S, extension at 72°C for 30S, a total...
Embodiment 3
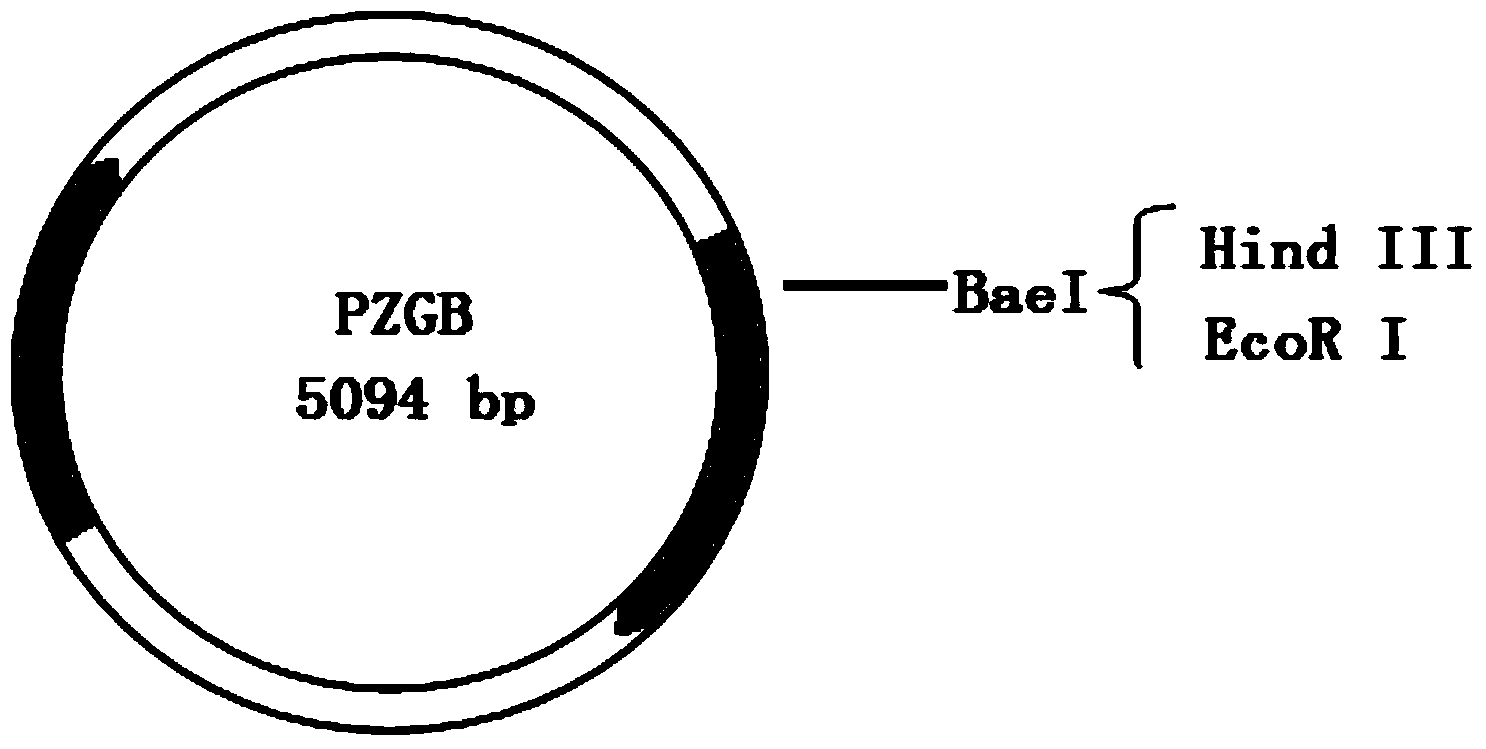
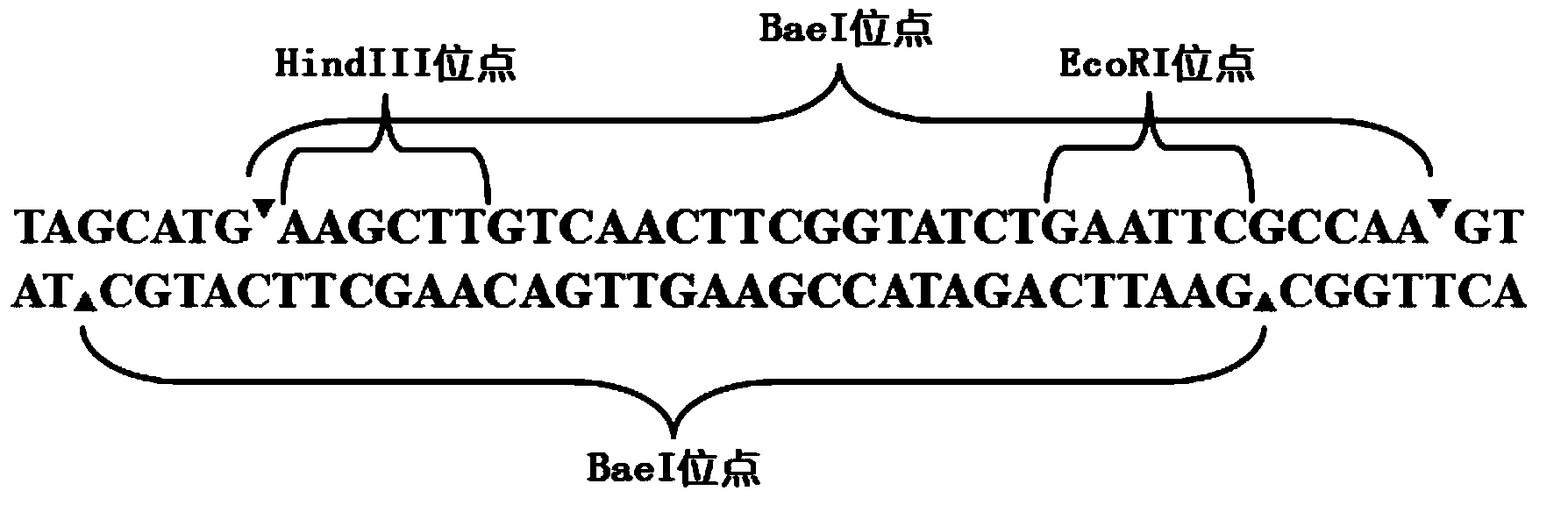
[0115] In Example 3, 32bp, 33bp, and 21bp heterologous nucleic acid sequences containing stop codons were respectively inserted into PZAB to verify the feasibility and sensitivity of the bleomycin reporter gene.
[0116] 1. Entrust a primer synthesis company to synthesize the following oligos:
[0117] 32+: CTAGAGATATCGTCGACCTGCAGAAGCTTCCGCGGTT, (Seq No. 10)
[0118] 32-:CGGAAGCTTCTGCAGGTCGACGATATCTCTAGCATAG,, (Seq No.11)
[0119] 33+:CCTAGAGATATCGTCGACCTGCAGAAGCTTCCGCGGTT, (Seq No.12)
[0120] 33-:CGGAAGCTTCTGCAGGTCGACGATATCTCTAGGCATAG, (Seq No. 13)
[0121] 21+: AAGCTGGATATCGTATAGCTTCGGTT, (Seq No. 14)
[0122] 21-: AAGCTATACGATATCCAGCTTCATAG, (Seq No. 15)
[0123] 1. Insert each length into the nucleic acid fragment oligo to construct the annealing system respectively. The Annealing system is:
[0124] oligonucleotide+100nmol / ml, oligonucleotide-100nmol / ml, 10× annealing buffer 1×, DEPC-water appropriate amount.
[0125] 10×Annealing buffer composition: 10mM Tris-HCL...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com