Bile salt hydrolase (BSH) mutant and use thereof
A technology of bile salt hydrolyzing enzyme and mutants, which is applied in the field of genetic engineering to achieve the effect of improving the hydrolysis ability
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0054] Example 1: Construction method of pET-20b(+)-dmsA-bsh
[0055] The specific operation is as follows:
[0056] Plasmid pUC57-Tat (Tat-dependent protein targeting in prokaryotes and chloroplasts) was used as a template to amplify the dmsA gene. The PCR conditions were: pre-denaturation at 95°C for 10 minutes; 30 cycles at 98°C for 10s, 55°C for 30s, and 72°C for 20s; and extension at 72°C for 10 minutes. At the same time, using the genomic DNA of L. plantarumBBE7 as a template, PCR amplified (with an extension time of 1 min) the bsh gene without a stop codon, the two fragments were gel-recovered, and the two fragments were used as templates in equimolar amounts to obtain Fusion fragment dmsA-bsh.
[0057] 2) Carry out double digestion with NdeI and XhoI, and connect with the correspondingly digested plasmid pET-20b(+), to obtain the recombinant plasmid pET-20b(+)-dmsA-bsh. The recombinant plasmid was transformed into E.coliJM109 competent cells, and positive recombinant...
Embodiment 2
[0058] Example 2: Alignment Analysis of Amino Acid Sequences
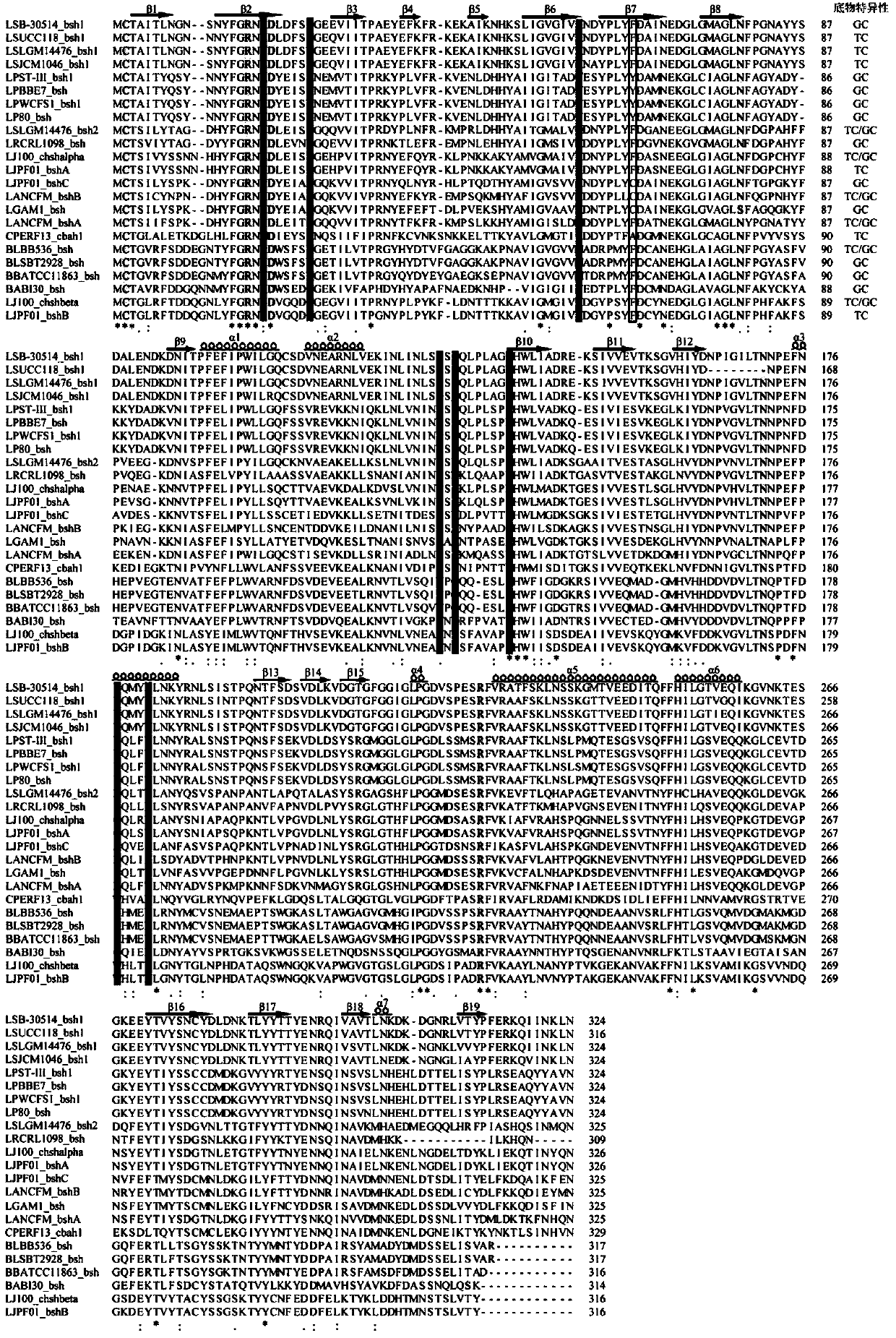
[0059] The amino acid sequences of BSH with known substrate specificities were aligned and analyzed using the online tool ClustalW2 (http: / / www.ebi.ac.uk / Tools / msa / clustalw2 / ). The bile salt hydrolase genes for comparative analysis include the following 23 species, the specific amino acid sequences are shown in figure 1 :
[0060] LSB-30514_bsh1, L. salivariusB-30514BSH1 (AFP87505.1);
[0061] LSUCC118_bsh1, L. salivariusUCC118BSH1 (ACL98201.1); LSLGM14476_bsh1, L.
[0062] salivariusLGM14476BSH1(ACL98197.1);LSLGM14476_bsh2,L.salivariusLGM14476(ACL98205.1);
[0063] LSJCM1046_bsh1.L.salivariusJCM1046BSH1(ACL98194.1);
[0064] LPST-III-bsh1, L.plantarum subsp.plantarumST-IIIBSH1 (ADO00098.1); LPBBE7_bsh, L.
[0065] plantarum BBE7BSH;
[0066] LPWCFS1_bsh1,L.plantarumWCFS1(CCC80500.1);
[0067] LP80_bsh,L.plantarum80(AAB24746.1);
[0068] LRCRL1098_bsh,L.reuteriCRL1098(ACH81023.1);
[0069] LJ100_chshalpha,...
Embodiment 3
[0084] Example 3: Homologous structure modeling and superposition
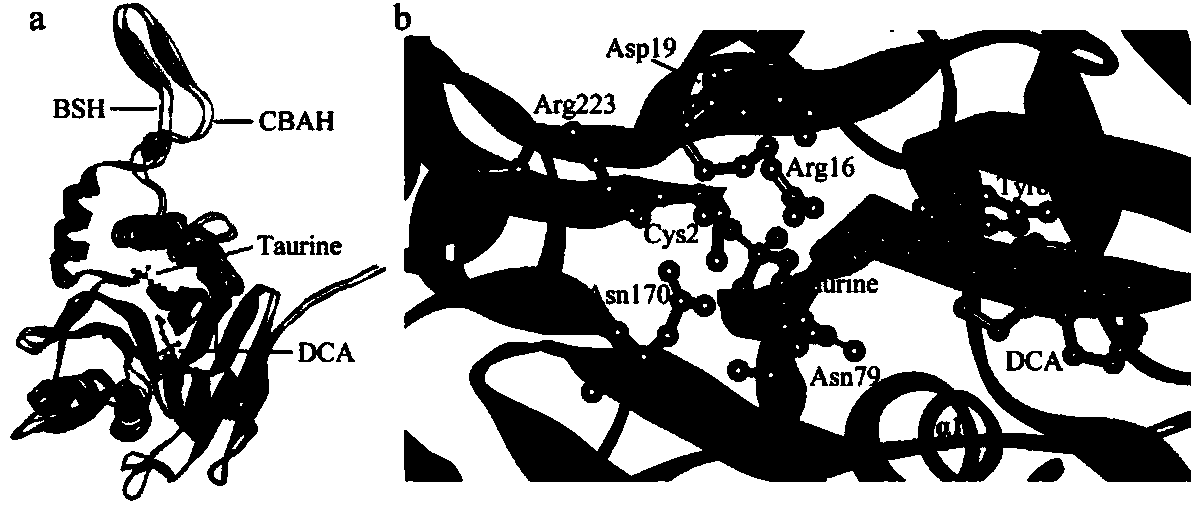
[0085] The homology simulation of the 3-D structure of L. plantarumBBE7BSH was performed using the online software SwissModel, and the template used for homology modeling was the mutant C2a (PDBID: 2rf8B) of C. perfringens13CBAH. The homology of the two sequences is 37.69%. The estimated absolute model quality (QMEANZ-Score) and the root mean square deviation (Root-Mean-SquareDeviation, RMSD) of the α-carbon atoms of the BSH model after simulation were -3.61 and Superimpose this model with the complex structure of CBAH and substrate (PDBcode: 2BJF, 2BJG) (RMSD is ), obtained the structure of the complex of L.plantarumBBE7BSH and deoxycholic acid (DCA), taurine ( figure 2 a). figure 2 Superposition of L.plantarumBBE7BSH and C.perfringens13CBAH structures in a, dark gray is BSH, light gray is CBAH; figure 2 b depicts the conserved amino acids (Cys2, Arg16, Asp19, Asn79, Asn170, and Arg223) in L. plantar...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com