Fluorescence detection method for copper ion based on click chemistry
A technology of fluorescence detection and click chemistry, which is applied in the field of chemical analysis and detection, can solve the problems of low sensitivity and cumbersome detection steps, and achieve the effects of strong anti-interference ability, reducing the possibility of false positive signals, and mild reaction conditions
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
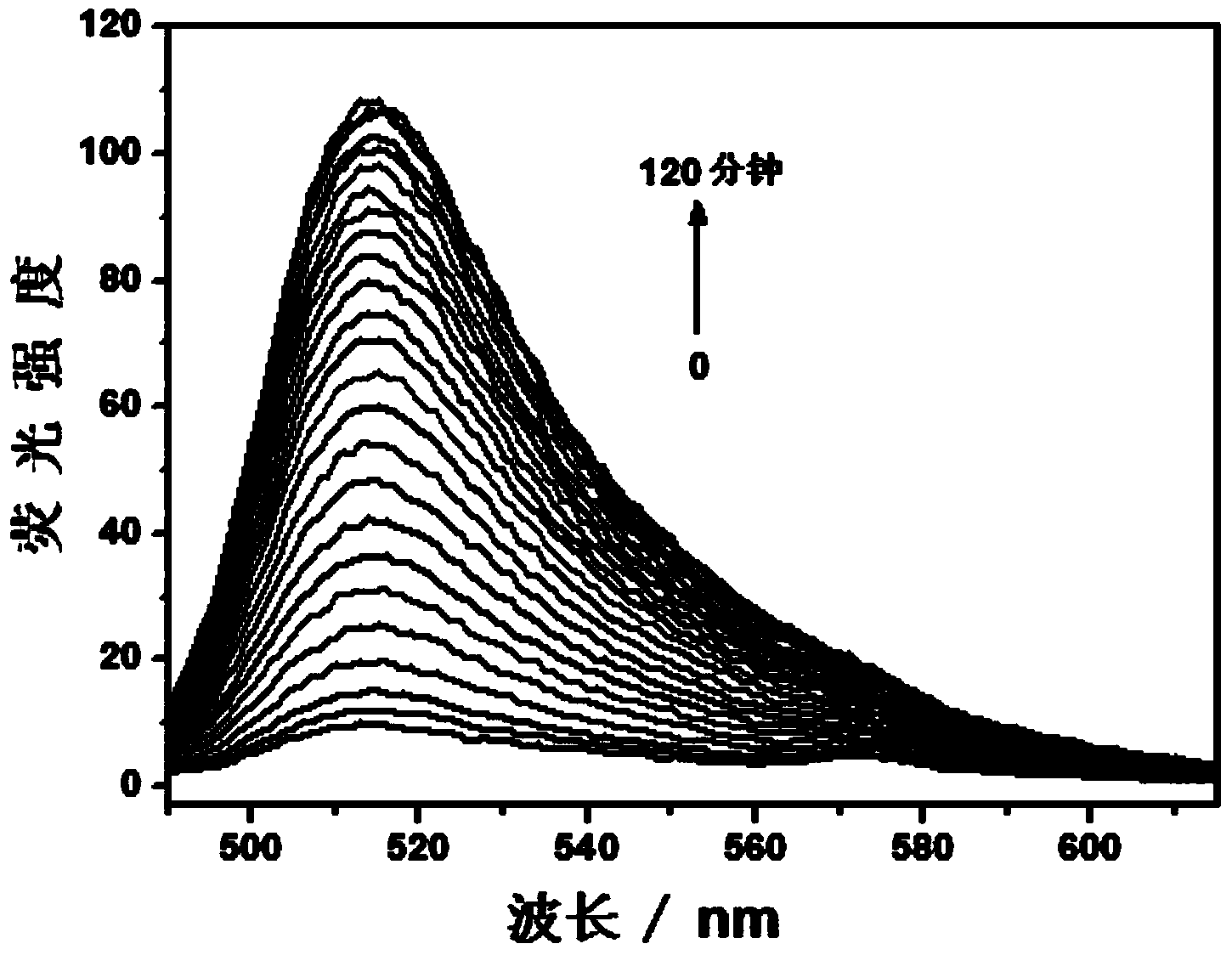
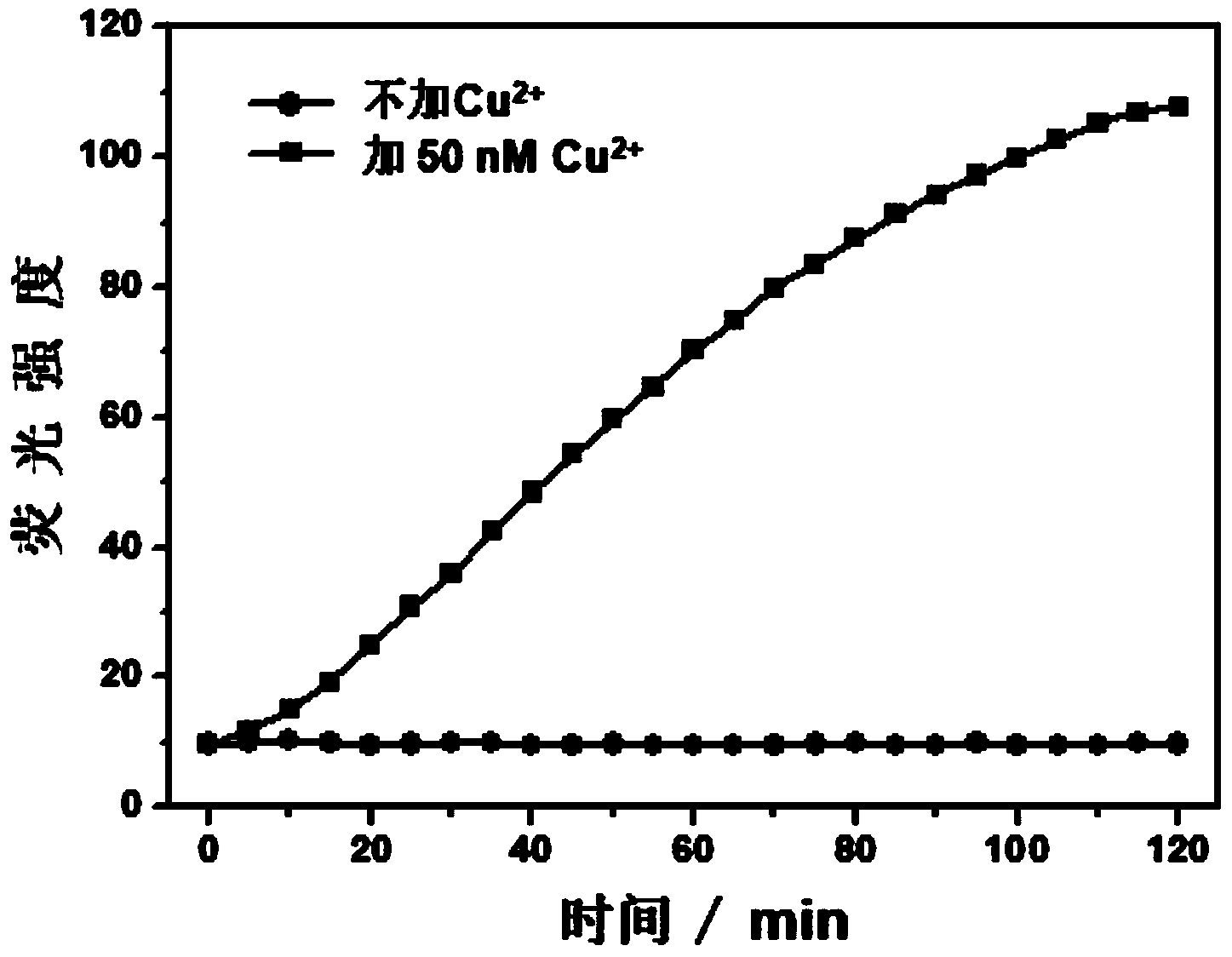
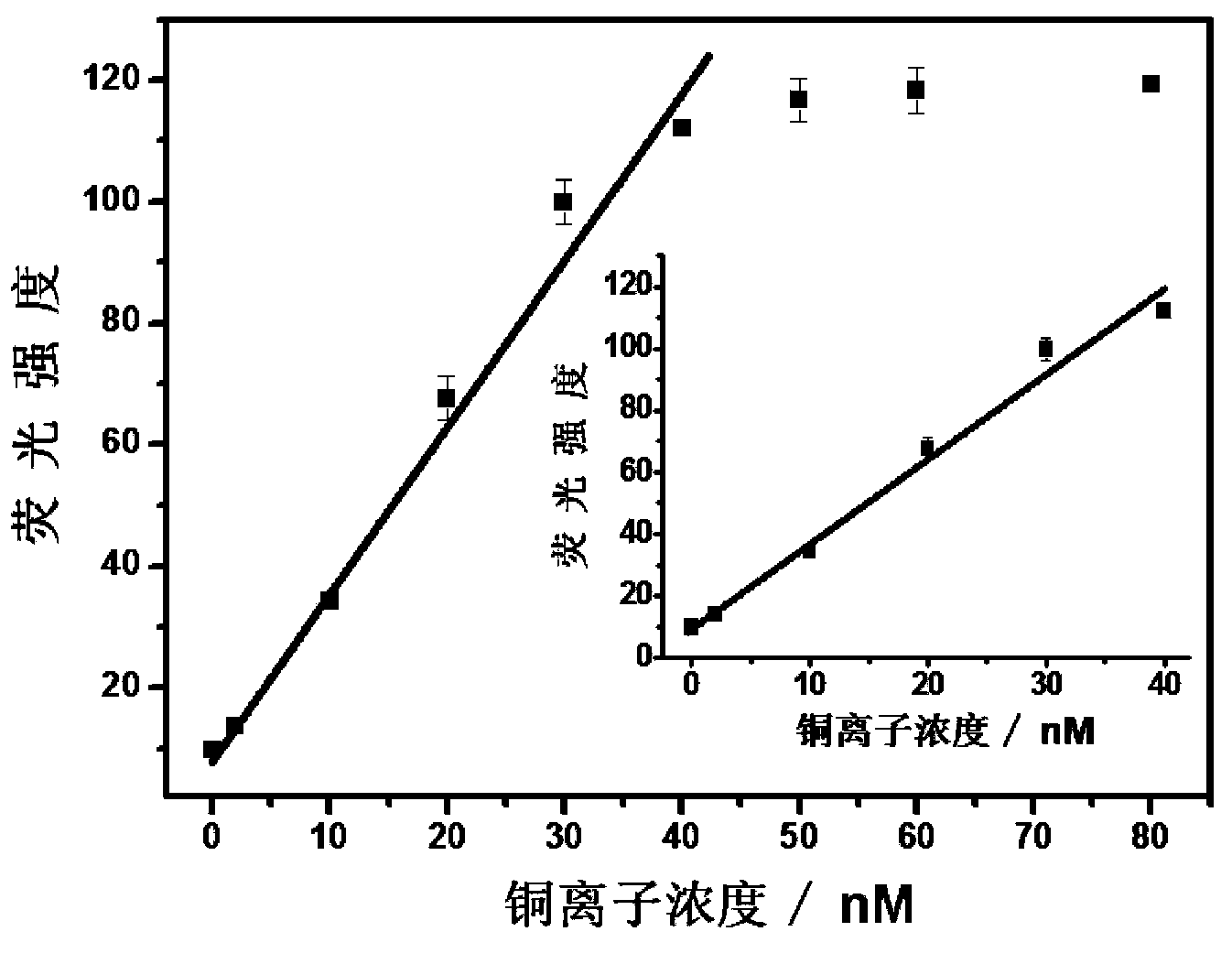
[0042] Step 1. The short single-stranded nucleic acid modified with an azide group, as shown in SEQ ID No: 1 (5'-CTA AAT TCC AA-azide-3'), was modified with a short alkynyl group. Single-stranded nucleic acid, as shown in SEQ ID No: 5 (5'-alkynyl-GAA ACT GAT AG-3'), and nucleic acid as ligation template, such as SEQ ID No: 9 (5'-FAM-TCG CTA TCA GTT TCT TGG AAT TTA GCG A-Dabcyl-3') were respectively dissolved in 20 mM Tris-HCl (pH 7.4) buffer solution to prepare nucleic acid solutions with a concentration of 6 μM;
[0043] Step 2. Mix the short single-stranded nucleic acid solution modified with azide group, the short single-stranded nucleic acid solution modified with alkynyl group, and the nucleic acid solution as a connection template in a ratio of 3:3:1 to form a mixed solution. solution, and then add 500 μM ascorbic acid, water and 20 mM Tris-HCl (pH 7.4) buffer solution to the mixed solution to form a detection system, in which the short single-stranded nucleic acid solut...
Embodiment 2
[0048] Step 1. The short single-stranded nucleic acid modified with an azide group, as shown in SEQ ID No: 1 (5'-CTA AAT TCC AA-azide-3'), was modified with a short alkynyl group. Single-stranded nucleic acid, as shown in SEQ ID No: 5 (5'-alkynyl-GAA ACT GAT AG-3'), and nucleic acid as ligation template, such as SEQ ID No: 9 (5'-FAM-TCG CTA TCA GTT TCT TGG AAT TTA GCG A-Dabcyl-3') were respectively dissolved in 20 mM Tris-HCl (pH 7.4) buffer solution to prepare nucleic acid solutions with a concentration of 6 μM;
[0049] Step 2. Mix the short single-stranded nucleic acid solution modified with azide group, the short single-stranded nucleic acid solution modified with alkynyl group, and the nucleic acid solution as a connection template in a ratio of 3:3:1 to form a mixed solution. solution, and then add 500 μM ascorbic acid, water and 20 mM Tris-HCl (pH 7.4) buffer solution to the mixed solution to form a detection system, in which the short single-stranded nucleic acid solut...
Embodiment 3
[0055] Step 1. The short single-stranded nucleic acid modified with an azide group, as shown in SEQ ID No: 1 (5'-CTA AAT TCC AA-azide-3'), was modified with a short alkynyl group. Single-stranded nucleic acid, as shown in SEQ ID No: 5 (5'-alkynyl-GAA ACT GAT AG-3'), and nucleic acid as ligation template, such as SEQ ID No: 9 (5'-FAM-TCG CTA TCA GTT TCT TGG AAT TTA GCG A-Dabcyl-3') were respectively dissolved in 20 mM Tris-HCl (pH 7.4) buffer solution to prepare nucleic acid solutions with a concentration of 6 μM;
[0056] Step 2. Mix the short single-stranded nucleic acid solution modified with azide group, the short single-stranded nucleic acid solution modified with alkynyl group, and the nucleic acid solution as a connection template in a ratio of 3:3:1 to form a mixed solution. solution, and then add 500 μM ascorbic acid, water and 20 mM Tris-HCl (pH 7.4) buffer solution to the mixed solution to form a detection system, in which the short single-stranded nucleic acid solut...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com