Thyroid Hormone Receptor α Gene as Genetic Marker for Goat Growth Traits and Its Application
A growth trait, α gene technology, applied in the application, genetic engineering, plant genetic improvement and other directions, can solve problems such as unseen TRα gene polymorphism
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
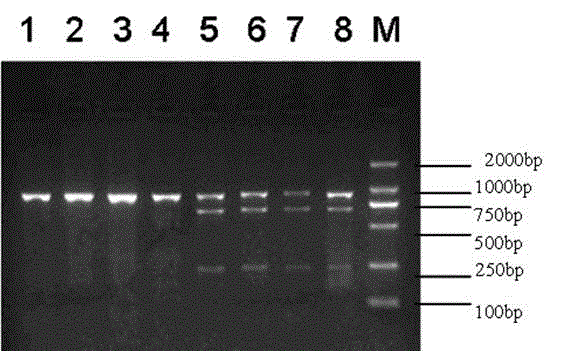
[0019] Example 1: Obtaining and Polymorphism Detection of Goat TRα Gene Fragment
[0020] Utilizing the NCBI database information, the bovine and sheep TRα genes were compared online, and primers were designed to amplify the sixth intron of the goat TRα gene in the conserved region of exons 6 and 7. Forward primer: 5'GCCTTCAGCGAGTTTACCAA3', reverse primer: 5'CTTCCGTGTCATCCAGGTTA3'.
[0021] Select 6 genomic DNA samples (3 Nanjiang yellow sheep and 3 Tibetan goats each), use their genomic DNA as a template, and use the above-mentioned primers to carry out PCR reaction. Reaction system: 2×MasterMixTaq enzyme 25 μL, cDNA 4 μL, upstream primer (10 μM) 1.5 μL, downstream primer (10 μM) 1.5 μL, ddH2O 18 μL. PCR reaction program: pre-denaturation at 95°C for 5 min; then denaturation at 94°C for 30 s, annealing at 61.2°C for 30 s, extension at 72°C for 45 s, and 35 cycles; finally, extension at 72°C for 10 min. Take 4 μL of the product, detect it by agarose gel electrophoresis, and ...
Embodiment 2
[0022] Example 2: Polymorphic distribution of molecular markers in different goat populations
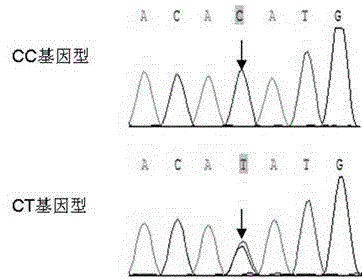
[0023] The detection found that the C230T loci had two genotypes CC and CT in the three breeds of Nanjiang yellow sheep, Inner Mongolia cashmere goat and Tibetan goat, and the CC genotype was the dominant genotype. This locus is in the Hardy-Weinberg equilibrium state in the three breeds, and it is in low-density polymorphism (PIC<0.25) in southern Xinjiang gazelle and Inner Mongolia cashmere goat, and in moderate polymorphism (0.25< PIC<0.5). As shown in the following table:
[0024] Table 1 The genotype (allele) frequency and genetic characteristics of TRα gene C230T locus in three goat breeds
[0025]
Embodiment 3
[0026] Example 3: Application of Molecular Markers in Production Traits
[0027] In order to establish the relationship between molecular markers of TRα gene and goat phenotype, 124 Nanjiang yellow sheep (Nanjiang Yellow Goat Breeding Farm, Nanjiang County, Sichuan Province) were selected as experimental materials. The test population was detected by the polymorphism detection method established in Example 1. The GLM process of SASv8.0 software was used to analyze the correlation between the C230T mutation site and the measured values of growth traits of Nanjiang Yellow sheep, and the results were expressed in LSM±standard error.
[0028] For the population of Nanjiang yellow sheep (124 individuals), the linear model was used to analyze the growth traits such as body weight, body length, body height, bust and newborn weight of different genotypes at the C230T site of TRα gene and different growth stages. Mainly: Y ijkl =μ ijkl +S i +B j +G k +S i ×B j +e ijkl , wher...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com