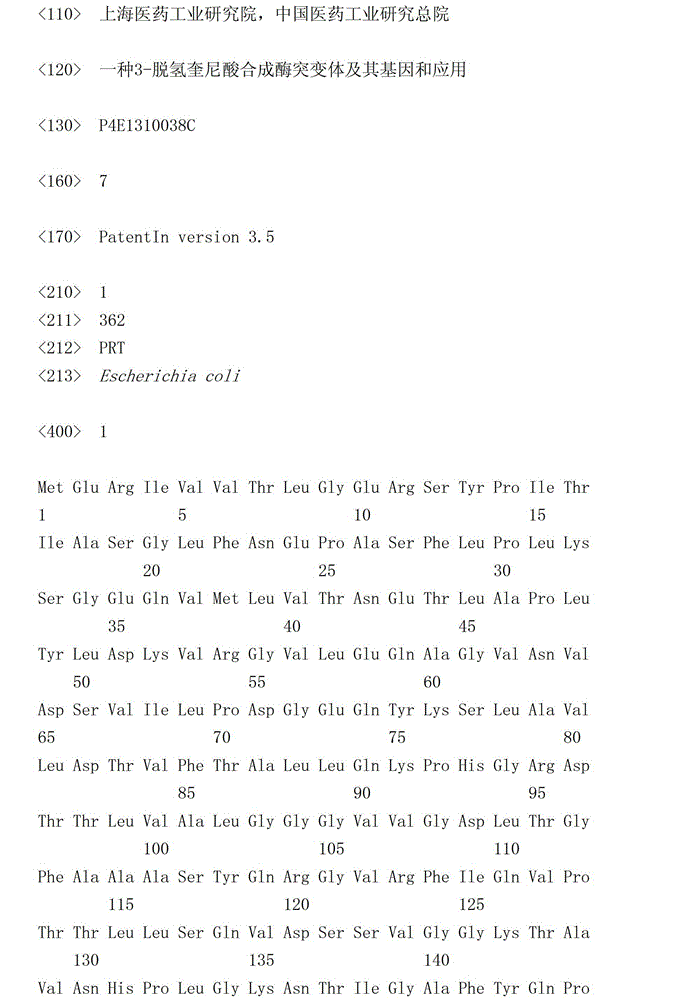A kind of 3-dehydroquinic acid synthase mutant and its gene and application
A technology for synthesizing enzymes and quinic acid, which is applied in the fields of genetic engineering and enzyme engineering, can solve the problems of insufficient activity and low yield of shikimic acid, and achieve the effects of improving enzyme activity, increasing activity, and increasing yield
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 13
[0030] The construction of the mutant library of embodiment 13-dehydroquinic acid synthase gene
[0031](1) Design primers for error-prone PCR amplification of 3-dehydroquinate synthase gene. The sequence of the upstream primer B1 (as shown in SEQ ID No: 2) is 5'-AAAACCATGGAGAGGATTGTCGTTACTCTCGG-3', and the downstream primer B2 ( As shown in SEQ ID No: 3), the sequence is 5'-GACAAAGCTTACGCTGATTGACAATCGGCAAT-3', which respectively contain NcoI and HindIII restriction sites.
[0032] (2) Using the genome of Escherichia coli W3110 (purchased from E.coliGeneticStockCenter, YaleUniversity, Yale University E.coli Collection Center) as a template, use primers B1 and B2 to carry out PCR amplification under the catalysis of TaqDNA polymerase to obtain 3-de Hydroquinic acid synthase gene fragment, the gene fragment obtained by the first round of error-prone PCR was recovered from agarose gel, and the second round of error-prone PCR was performed using it as a template. The PCR reaction...
Embodiment 23
[0034] Example 23 - Screening of a library of mutants of dehydroquinic acid synthase gene
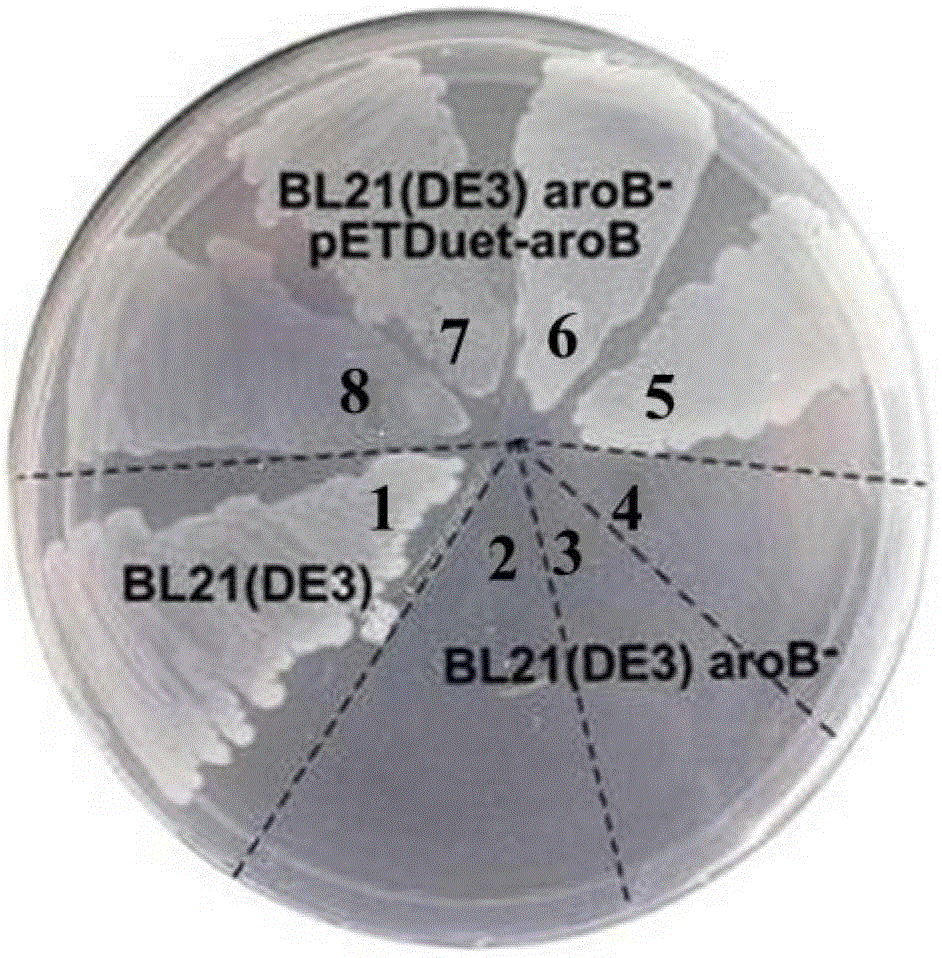
[0035] (1) Construction of 3-dehydroquinic acid synthase gene-deficient strain
[0036] The plasmid pKD46 expressing Red recombinase (purchased from Yale University Escherichia coli Collection Center, the plasmid information is disclosed in the literature BabaT, AraT, HasegawaM, et al.ConstructionofEscherichiacoliK-12in-frame, single-geneknockoutmutants:theKeiocollectionMolSystBiol2006:2-8) Introduce E.coliBL21(DE3), and select transformants on LB solid plates containing 100 mg / mL ampicillin (cultivation temperature is 30°C). Inoculate the transformant into LB liquid medium, shake culture at 200r / min at 30°C, when the cell OD 600 When it reaches 0.2, add 1mML-arabinose to induce the expression of Red recombinase, the induction time is not less than 1h, OD 600 Harvest the cells at about 0.6 to prepare competent cells. The fragments for homologous recombination were introduced into com...
Embodiment 33
[0039] Example 33 - Expression of dehydroquinic acid synthase gene mutants
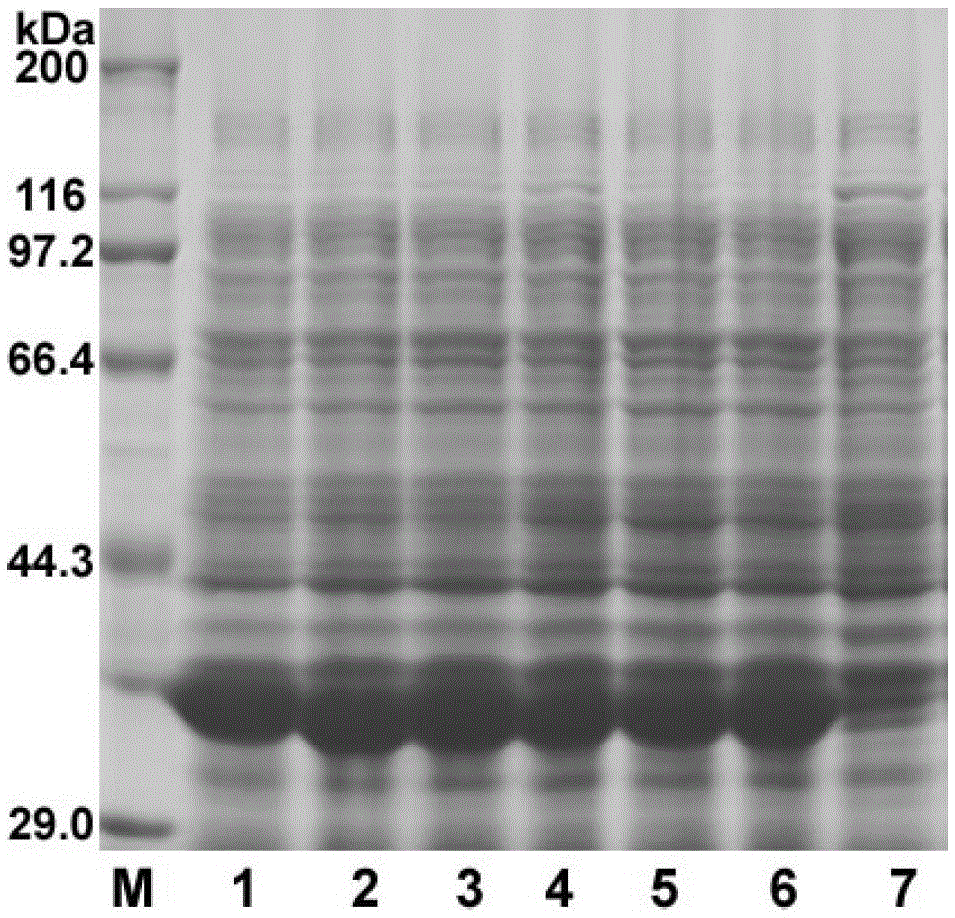
[0040] Inoculate the mutant strains grown on the M9 solid medium in Example 2 into a 250ml Erlenmeyer flask containing 20ml of seed medium, and after overnight shaking at 37°C, inoculate into fresh LB medium according to 1% inoculum size, 37 Cultivate to OD 600 At about 0.6, add IPTG to a final concentration of 1 mM, continue to induce culture for 4 hours, centrifuge the bacterial solution at 12,000 r / min for 5 minutes, discard the supernatant, and use the bacterial cells for SDS-PAGE detection. The result is as figure 2 As shown, compared with the No. 7 host strain, No. 1-6 strains expressed the target protein of 38 kDa, and the expression level was higher.
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com