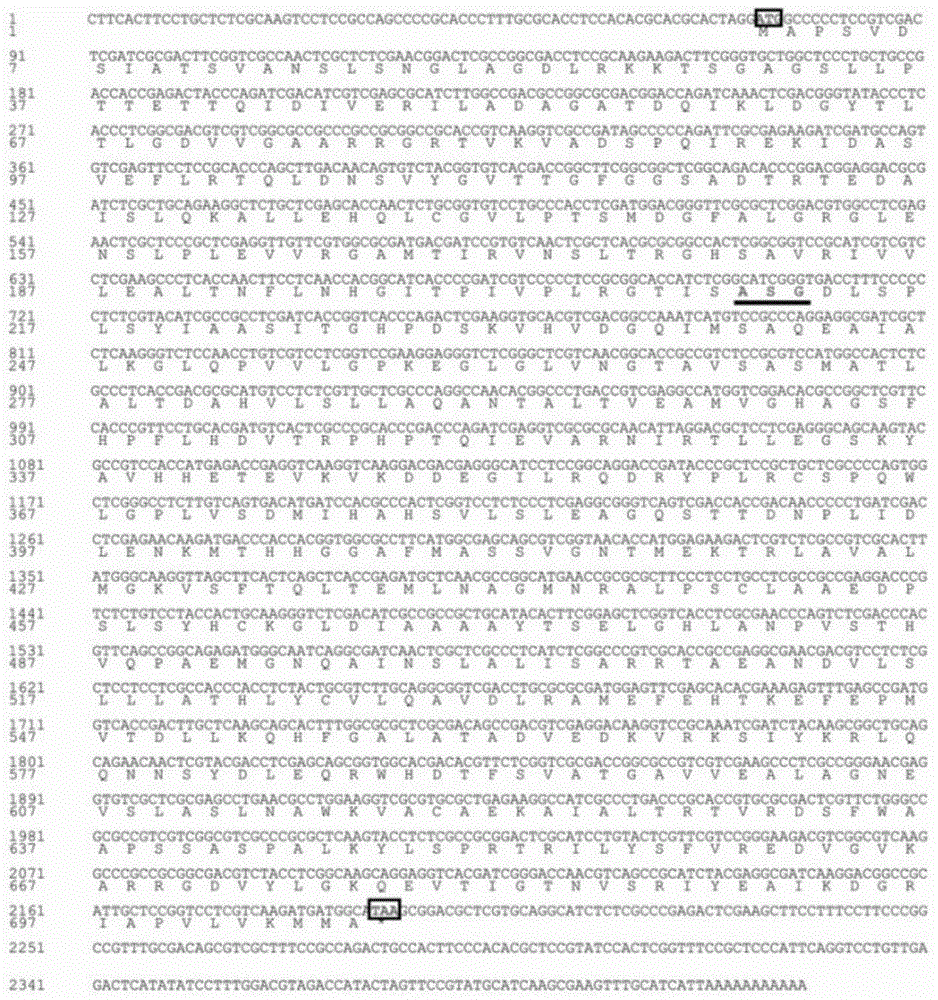
Rhodotorula glutinis phenylalanine deaminase gene and application thereof
A technology of phenylalanine deaminase and gene, which is applied in Rhodotorula viscosus phenylalanine deaminase gene and its application field, can solve the problems of high GC content and difficulty in reverse transcription, and improve enzyme activity Effect
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
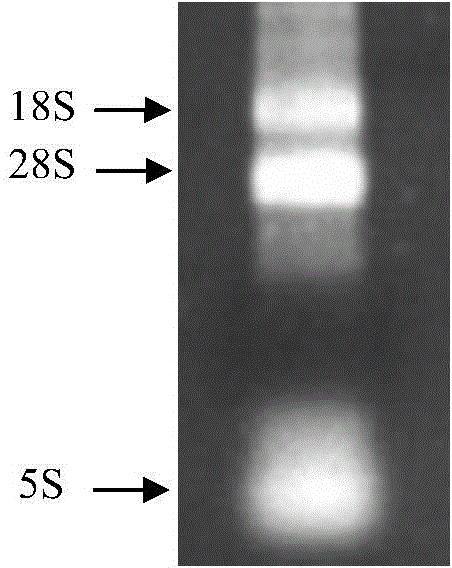
[0019] Example 1 Extraction of Rhodotorula viscosus total RNA
[0020] (1) Using the strain Rhodotorula glutinis (Rhodotorula glutinis) CCTCC NO:M2011490 used for industrial production as the starting strain, pick a single colony from the Rhodotorula glutinis plate and put it into 5 mL of seed medium, culture it at 30°C for 24 hours, and take 50 μL of Put the seed solution into a 250mL Erlenmeyer flask filled with 50mL medium, and culture at 30°C for 18-20h to OD 600 =1.0, collect the bacterial cells by centrifugation at 1500 g for 5 min, wash the bacterial cells twice with cold water, and discard the supernatant.
[0021] (2) Resuspend the cells with 400 μL of TES (10mM Tris-HCl, 10mM EDTA, 0.5% SDS, pH7.5) buffer, add 400 μL of acidic phenol, shake vigorously for 10 sec, incubate at 65°C for 1 hour, and vortex from time to time The device oscillates briefly.
[0022] (3) Incubate on ice for 5 minutes, and centrifuge at 12000g for 5 minutes at 4°C.
[0023] (4) Transfer th...
Embodiment 2
[0028] Example 2 Reverse transcription to obtain cDNA
[0029] (1) Add 50ng total RNA and 1μL 5'-CDS(5'-(T) to a 0.5mL centrifuge tube 25 VN (N = A, C, G, T; V = A, G, C)-3') and 1 μL SMARTer II A Oligonucleotide (5'-AAGCAGTGGTATCAACGCAGAGTACGGGGG-3'), add water to 4.75 μL and mix well.
[0030] (2) Incubate at 72° C. for 3 minutes, then incubate at 42° C. for 2 minutes, and centrifuge at 1500 g for 10 sec.
[0031] (3) Add 2μL of 5×buffer (250Mm Tris-HCl, 375mM KCl, 30mM MgCl 2 ), 1 μL DTT (20mM), 1 μL dNTP (10mM), 0.25 μL RNase inhibitor and 1 μL SMARTer TM For reverse transcriptase, incubate at 42°C for 90 minutes, denature at 70°C for 10 minutes, cool to room temperature and add 20 μL sterile water to obtain cDNA.
Embodiment 3
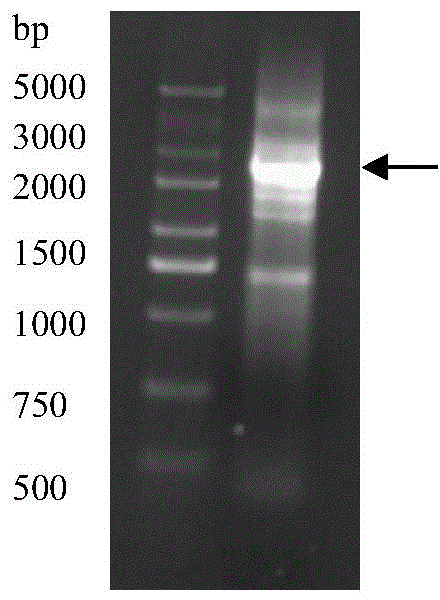
[0032] Example 3 Obtaining the sequence of the 5' end of pal
[0033] (1) Add 1 μL cDNA, 5 μL Mix primer (5'-CTAATACGACTCACTATAGGGCAAGCAGTGGTATCAACGCAGAGT-3'; 5'-CTAATACGACTCACTATAGGGC-3'), 1 μL GSP-specific primer (5'-GGTGATGCCG TGGTTG AGGAAGTT-3') to a 0.25 mL centrifuge tube , 5μL 10×buffer, 5μL dNTP (2mM), 0.5μL pfu DNA polymerase, 3μL MgSO 4 (25mM), add deionized water to 50μL.
[0034] (2) Perform PCR amplification, PCR amplification conditions: pre-denaturation at 94°C for 4 min, denaturation at 94°C for 1 min, annealing at 58°C for 30 sec, extension at 72°C for 60 sec, 25 cycles, and detection by agarose gel electrophoresis.
[0035] (3) Add 5 μL of PCR product, 1 μL of 10×Taq buffer, 1 μL of dATP (2mM), 1 μL of Taq enzyme (5U / μL) into a 0.25 mL centrifuge tube, add water to 10 μL, react at 70°C for 30 minutes, extract with phenolform, and precipitate with ethanol , aseptic operation typhoon for 15 minutes until the alcohol evaporates completely, and resuspended with...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com