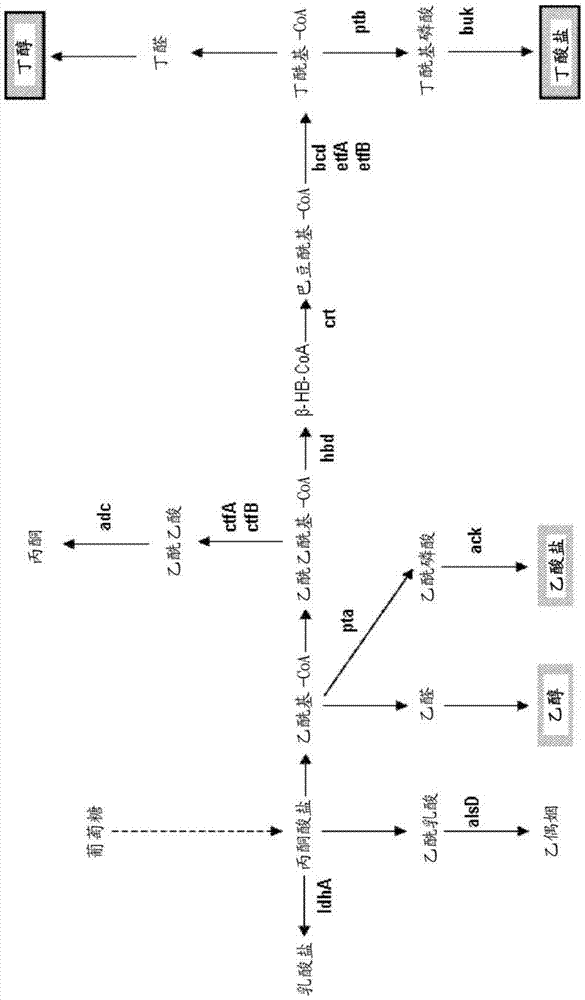
A method for producing butanol
A technology of n-butanol and species, applied in the field of butanol production
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0100] Example 1: Plasmid construction and chromosome modification scheme
[0101] The protocol described below was used to construct a genetically distinct C. acetobutylicum strain ATCC824 lacking the rex gene or the pta-ack gene as mentioned in the examples below.
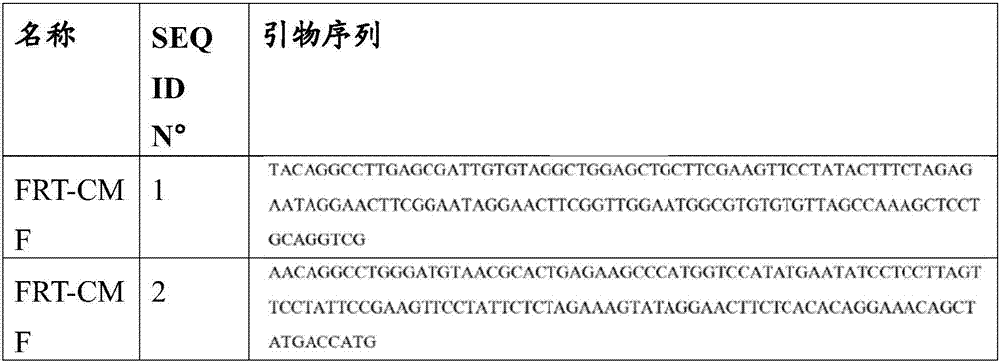
[0102] 1.1 Construction of pUC18-FRT-Pptb-catP vector
[0103] This plasmid contains a novel CM / TH antibiotic marker "catP" that is functional in Clostridium and flanked by two FRT sites, and is useful for constructing a replacement cassette. The "catP" gene is described in patent application WO2009 / 137778. The "catP" sequence flanked by two FRT sites (two StuI site) and under the control of the Clostridium acetobutylicum phosphotransbutyrylase (ptb) promoter. The PCR product was phosphorylated by T4 polynucleotide kinase and cloned into SmaI digested pUC18 to generate the pUC18-FRT-Pptb-catP plasmid.
[0104]
[0105] 1.2 Construction of pSOS95-MLSr-upp vector
[0106] The upp gene with its own ribosome ...
Embodiment 2
[0114] Example 2: Deletion of the CAC_2713 gene encoding the redox-sensing transcriptional repressor REX of Clostridium acetobutylicum ATCC824
[0115] To delete the rex regulator (CAC_2713), Protocol 1 was used except that the primer pairs Rex 1-Rex 2 and Rex3-Rex 4 were used to amplify the 1135 bp upstream and 1138 bp downstream homology regions of the rex locus, respectively. Both primers Rex 1 and Rex4 introduce NotI sites, while primers Rex2 and Rex3 have complementary regions that introduce NruI sites. The DNA fragments Rex1-Rex2 and Rex3-Rex4 were joined in a PCR fusion experiment using primers Rex1 and Rex4, and the resulting fragment was cloned into pCR4-TOPO-Blunt (Invitrogen) to generate pTOPO : Rex. At the unique NruI site of pTOPO:Rex, an antibiotic resistance catP gene (Cm / Tm) flanked by FRT sequences was introduced from the 1230 bp StuI fragment of pUC18-FRT-Pptb-catP. The Rex deletion cassette obtained after digestion of the resulting plasmid with NotI was cl...
Embodiment 3
[0120] Example 3: Deletion of the CAC_2713 gene encoding the C. acetobutylicum ATCC824Δcac15ΔuppΔbukΔldhA redox-sensing transcriptional repressor REX
[0121] The Clostridium acetobutylicum Δcac15 Δupp Δbuk ΔldhA strain disclosed in patent application WO2008 / 052596 was transformed by electroporation using the pSOS95-MLSr-upp-rex-catP plasmid described in Example 2. The genotypes of thiamphenicol-resistant and clarithromycin-sensitive clones were detected by PCR analysis (using primers Rex 0 and Rex 5 located outside the Rex deletion cassette). Isolation of △cac15△upp△buk△ldhA△rex-catP that has lost the pSOS95-MLSr-upp-rex-catP vector R strain. The thiamphenicol resistance of the above strains was eliminated according to protocol 1. The genotype of the Δcac15ΔuppΔbukΔldhAΔrex strain that had lost the pSOS95-MLSr-upp-flp plasmid was checked by PCR analysis using primers Rex 0 and Rex 5 .
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com