Ginseng identification method and special kit
A kit and ginseng technology, applied in the field of molecular biology, can solve problems such as unfavorable application and promotion, cumbersome fluorescent staining methods, and long testing time
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0032] Embodiment 1, the preparation and use method of the kit for identifying ginseng
[0033] 1. Design and synthesis of primer pairs for identification of ginseng
[0034] In this example, the general primer 18S rRNA (18SF: 5'-CAACCTGCTTGATCCTGCCAGT-3'18SR: 5'-CTGATCCTTTCTGCAGGTTCACCTA-3') was selected to amplify the samples of Panax ginseng, Panax notoginseng, Panax notoginseng, Pearl ginseng and Panax notoginseng. Amplified, sequenced, and then homologously compared the obtained sequences, and designed a pair of specific primers F1 and F2 according to the variable region, as a pair of specific primers for identifying ginseng. The sequence is as follows:
[0035]Primer F1: 5'-ATAACAATACCGGGCTGATTC-3' (SEQ ID NO: 1);
[0036] Primer F2: 5'-GCCAGTTAAGGACAGGAG-3' (SEQ ID NO: 2).
[0037] 2. Assembly of kits for identification of ginseng
[0038] After the primers F1 and F2 designed and synthesized in step 1 are packaged separately, they are packaged in the same kit with s...
Embodiment 2
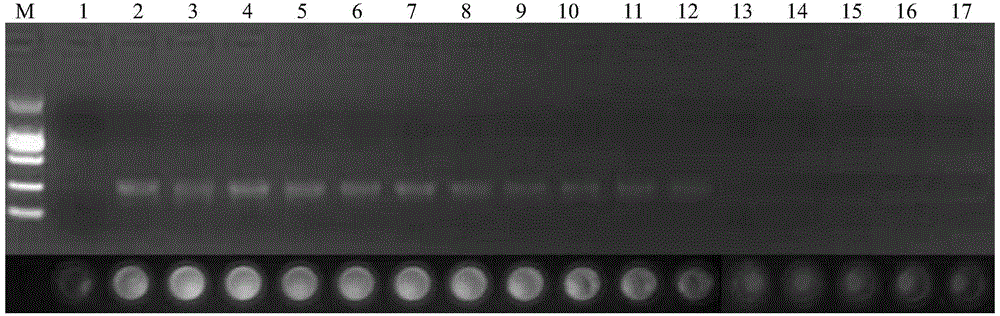
[0051] Embodiment 2, using the kit prepared in embodiment 1 to identify ginseng
[0052] Samples to be tested: 6 samples of Chinese herbal medicines shown in Table 1, ginseng, American ginseng, Panax notoginseng, bamboo ginseng, pearl ginseng, and Panax notoginseng.
[0053] 1. Extract genomic DNA from the sample to be tested
[0054] Select about 0.03 g of dry medicinal material without mildew, put it in a pulverizer, grind it, and pass it through a 40-mesh sieve. Transfer the powder to a 2.0 mL microcentrifuge tube, add 900 μL of sterilized CTAB extract (recipe: 2% (2g / 100ml) CTAB, 100mmol / L Tris-HCl pH=8.0, 20mmol / L EDTA, 1.4mol / L NaCl), 0.02g PVP 40000, 10μL β-mercaptoethanol, fully vortexed and mixed, 65 ℃ water bath for 1.5h-2h, during which, shake gently 2-3 times. After completion, take it out and cool to room temperature, add 900 μL of chloroform-isoamyl alcohol (volume ratio 24:1), shake and mix well, and centrifuge at 12000 g for 10 min. Take the supernatant, ad...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com