Molecular marker primer for detecting aneuploid hybrid progeny plants, and detection method of aneuploid hybrid progeny plants
A technology of molecular markers and aneuploidy, applied in the field of plant genetics and breeding in biotechnology, can solve the problems of restricting the wide application of emerging molecular karyotype analysis technology and high detection cost
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0035] 1. Preliminary detection of ploidy level of hybrid progeny plants
[0036] Take 50 mg of young leaves of progeny plants to be tested, put them on ice, add 1 mL of refrigerated nucleus isolation buffer, wherein, the nucleus isolation buffer is 45mM MgCl2 6H2O, 20mM MOPS, 30mM sodium citrate, 0.5% Triton X-100 , 1% PVP-10, pH 7.0; then quickly chopped into pieces with a single-sided blade for 60 seconds, the separation buffer containing nuclei was filtered through a gauze with a pore size of 40 μm, and a concentration of 50 μg / mL PI (Propidium Iodide, iodine) was added to the filtrate Propidium staining solution) solution, mixed evenly, wherein the PI solution contains RNase, the concentration of RNase in the PI solution is 50 μg / mL, then incubated in an ice bath for staining for 30min, and then used a flow cytometer (Partec, Germany) , model: CyFlow PA flow cytometer) for nuclear DNA content analysis.
[0037] The nuclei of both parents (ie, father and mother) were used...
Embodiment 2
[0084] 1. Preliminary detection of ploidy level of hybrid progeny plants
[0085] Same as Example 1.
[0086] 2. Extract the DNA of the father, mother and hybrid offspring
[0087] Same as Example 1.
[0088] 3. Primer screening
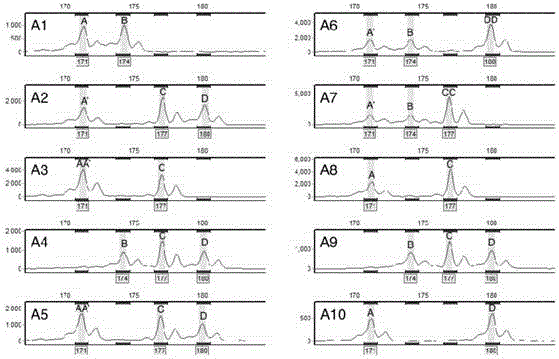
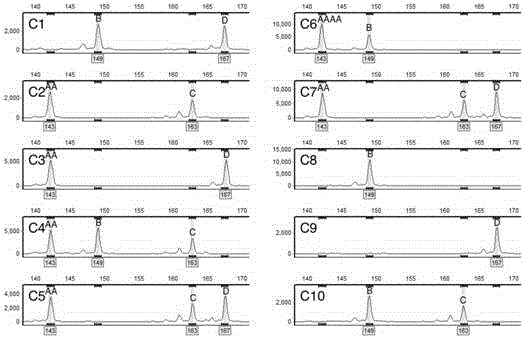
[0089] Based on the set marker requirements, through screening, the physical location of the marker site is close to the centromere, the parental differences and co-dominant segregation molecular marker primers, the primer PMGC-2607 located on chromosome VIII of poplar (see Table 3) is an example analysis, which is derived from the database of the International Poplar Genome Consortium (http: / / web.ornl.gov / sci / ipgc / ssr_resource.htm).
[0090] 4. PCR amplification and capillary fluorescence electrophoresis analysis
[0091] 4-1. Use PMGC-2607 primers to perform PCR amplification on the DNA of the hybrid parent and perform capillary fluorescence electrophoresis on the amplified product, wherein, the PCR reaction procedure and capillary fluorescence...
Embodiment 3
[0109] 1. Preliminary detection of ploidy level of hybrid progeny plants
[0110] Same as Example 1.
[0111] 2. Extract the DNA of the father, mother and hybrid offspring
[0112] Same as Example 1.
[0113] 3. Primer screening
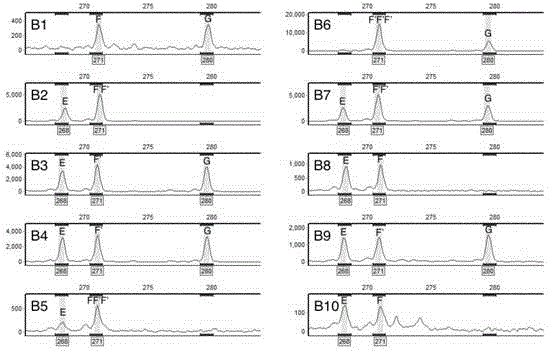
[0114] Based on the set marker requirements, through screening, the physical location of the marker site is close to the centromere, the parental differences and co-dominant segregation molecular marker primers, primer LG-10-1788 located on chromosome X of poplar (See Table 3) for example analysis, derived from the report of Yin et al. (2009).
[0115] Table 3 The SSR primers suitable for the identification of chromosome number VIII and X in the offspring of ‘Pulus maoxin’בYinzhong’
[0116]
[0117] 4. PCR amplification and capillary fluorescence electrophoresis analysis
[0118] 4-1. Use LG-10-1788 primers to perform PCR amplification on the DNA of the hybrid parent and perform capillary fluorescence electrophoresis on the amplified produc...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com