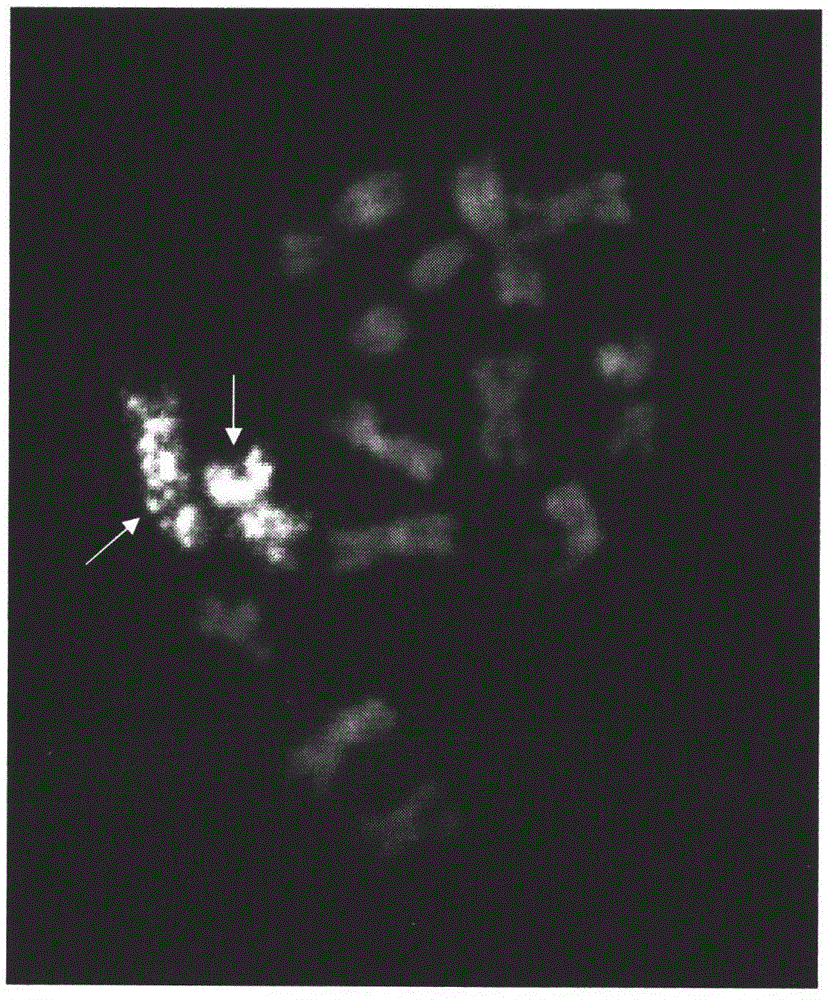
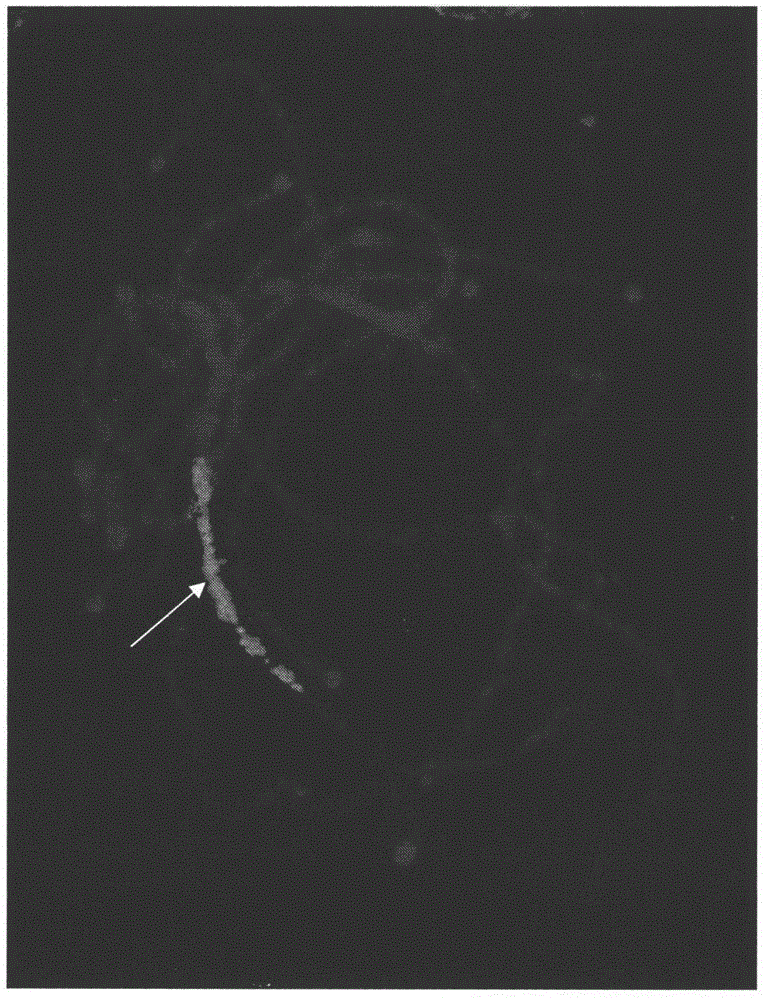
Preparation method and application of Cucumis sativus. L chromosome-6 probe
A chromosome and single-chromosome technology, applied in the field of plant biology, to achieve the effect of avoiding cumbersomeness and interference of repetitive sequences
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment Construction
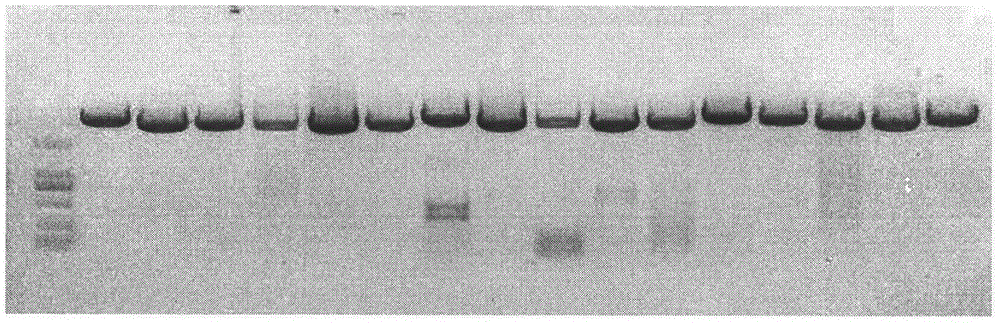
[0023] (1) Partial sequences were selected from the cucumber genome sequence at intervals of ~100kb, and repeated sequences were compared using RepeatMasker software (http: / / www.repeatmasker.org) and BLAST analysis, and sequences without repeated sequences were screened out.
[0024] (2) Use Primer Premier 5.0 software to design primers. The primer length is set to 20 bp, the Tm value is set to 55-65° C., the GC ratio is set to 40-60%, and the PCR product length is set to 2500-4000 bp. Primers with a Rating value above 75 and without hairpin structure, dimer structure, mismatch and dimer structure crossover were selected for repeat sequence alignment and BLAST analysis, and ideal primers without repeat sequences were screened out.
[0025] (3) Using cucumber genomic DNA as a template, amplify according to the PCR system. PCR reaction system:
[0026] 5×buffer: 4ul
[0027] dNTPs: 1.6ul
[0028] DNA: 1.2ul
[0029] Prime Star: 0.6ul
[0030] dd water: 12ul
[0031] Sense ...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com