Method for detecting activity of uracil-DNA glycosylase (UDG) based on fluorescence amplification strategy of label-free non-enzyme DNA machines
A technology of glycosylase activity and uracil, which is applied in the field of protein qualitative and quantitative detection, can solve the problems of sensitive protease reaction conditions, poor test result reproducibility, interference signal amplification ability, etc., to achieve low impact, improve sensitivity, low The effect of background signals
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0043] Example 1: Pure UDG activity detection
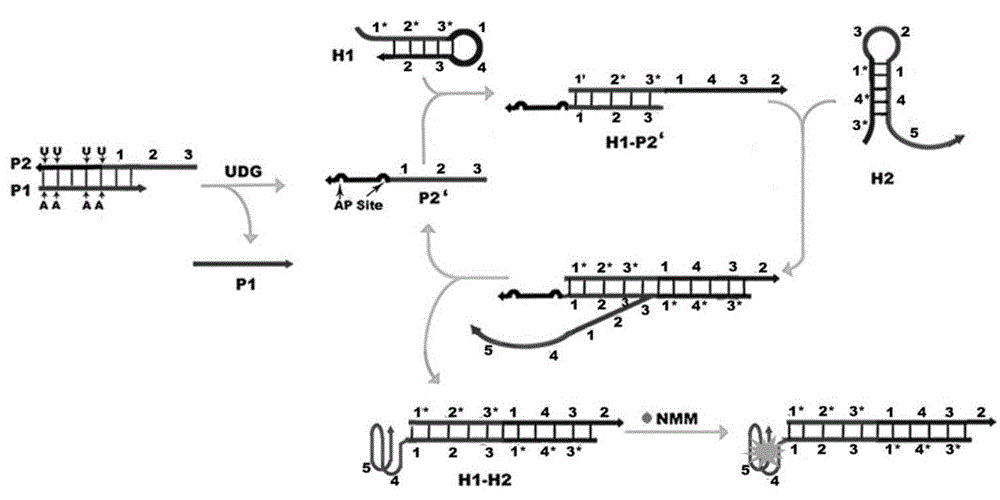
[0044] First design a double-stranded DNA (dsDNA) probe P1-P2 containing a uracil base and a priming sequence, wherein the P1 strand is an inhibitory strand, and its nucleotide sequence is as shown in SEQ ID NO.1 in the sequence listing; the P2 strand It is a chimeric conjugated strand containing a uracil DNA sequence and a priming sequence, and its nucleotide sequence is shown in SEQ ID NO.2 in the sequence table; the P1 strand and the P2 strand are partially complementary to form a double-stranded DNA (dsDNA) probe P1- P2;
[0045] P1: 5'-GAAATTCTTAAGTAGTCAG-3';
[0046] P2:5'-CGACATCTAACCTAGCTGACTACUUAAGAAUUTC-3'; (wherein, the part in italics is the triggering sequence)
[0047] According to the priming sequence of the P2 chain, two partially complementary hairpin probes H1 and H2 were designed for the construction of a label-free and enzyme-free DNA machine, and the G-quadruplex (G4) sequence was grafted on the end of the ...
Embodiment 2
[0052] Example 2: Detection of UDG activity in cervical cancer (Hela) cell lysate
[0053] Adopt detection method of the present invention to analyze and detect the UDG activity in Hela cell lysate, concrete steps are as follows:
[0054] Hela cell samples were pelleted by centrifugation (5 min, 3000 rpm, 4°C) and redispersed in lysis buffer (10 mM Tris-Hcl, pH 7.0) by sonication on ice. The mixed solution was then centrifuged at 12,000 rpm for 30 min at 4°C to remove insoluble substances. The supernatant was collected and filtered through a 0.45 μm filter membrane to prepare a crude Hela cell lysate.
[0055] Crude Hela cell lysates can be directly assayed for UDG activity without further processing. The detection method is the same as the pure UDG activity detection method in Example 1.
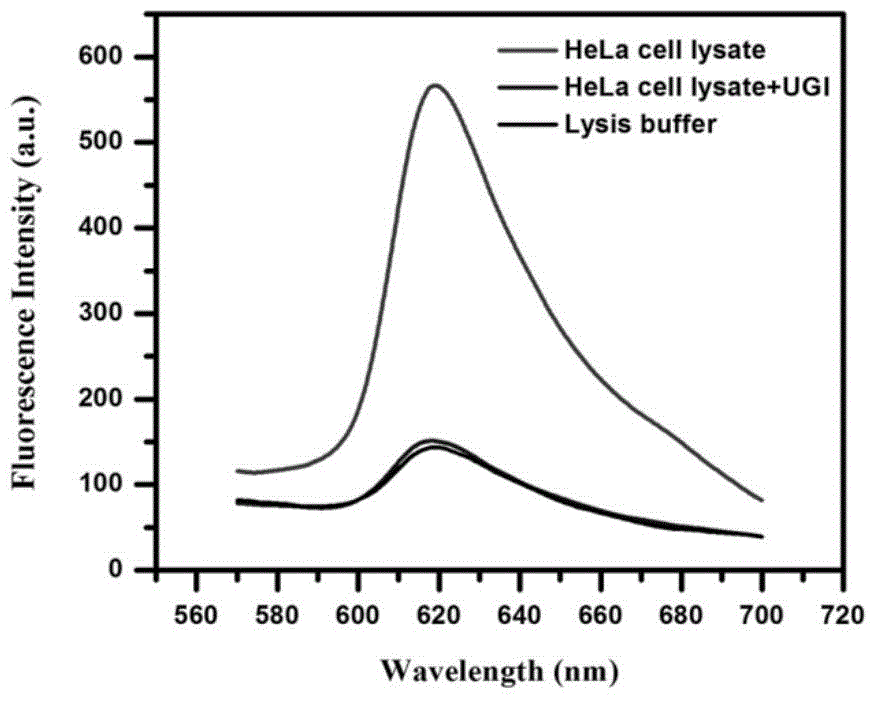
[0056] In order to verify the applicability of the detection method of the present invention in the detection of UDG activity, we also used the method of the present invention to analyze...
Embodiment 3
[0057] Example 3: Detection of activity inhibition of UDG
[0058] In order to detect the activity inhibition of UDG, the prepared dsDNA probes P1-P2 were mixed with 1 U / mL UDG and different concentrations of UGI, and incubated at 37°C for a certain period of time. Hairpins H1 and H2 were then added to the mixture to give a system with a final volume of 100 μL. At 37°C, the DNA machine performs amplification reactions for a certain period of time. Finally, NMM was added to the product and incubated at 37°C for 30min. All experiments were repeated three times.
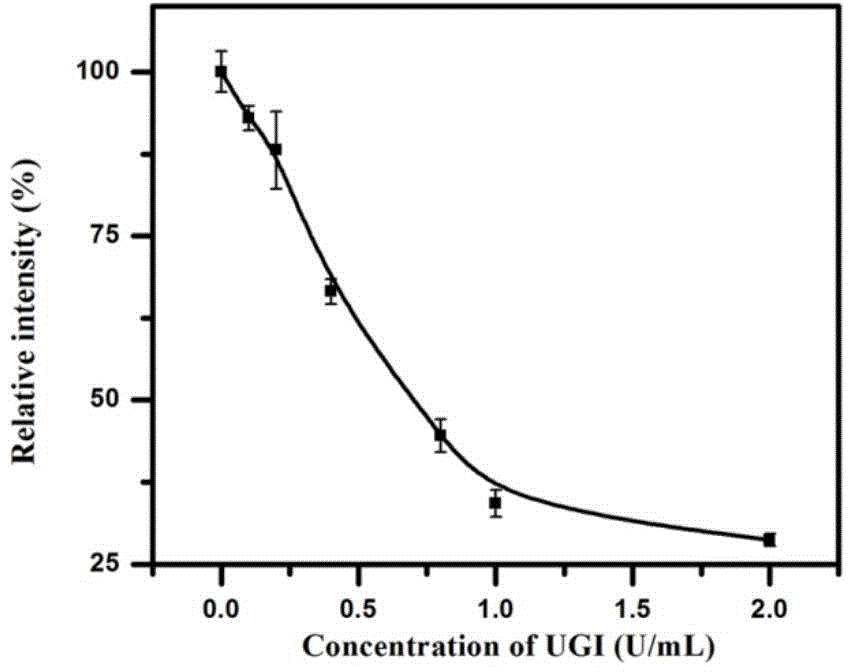
[0059] UGI was chosen as a model inhibitor. UGI can form a tight and physiologically irreversible complex with UDG at a molar ratio of 1:1, such as image 3 As shown, when UGI exists, the relative fluorescence intensity of the system decreases with the increase of UGI concentration. The results demonstrate that this strategy can be used to detect the activity inhibition of UDG.
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com