Molecular marker for identifying genotype of rice chalkiness major effect gene and application of molecular marker
A technology of molecular markers and major genes, applied in the field of plant biology, can solve the problems of difficulty in meeting breeding needs and high costs, and achieve the effect of meeting large-scale molecular breeding, moderate marker amplification fragments, and improving breeding efficiency
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0034] Example 1 Primer design and amplified fragment analysis of the functional marker Chalk5-T / C of rice chalky gene
[0035] 1. Primer design
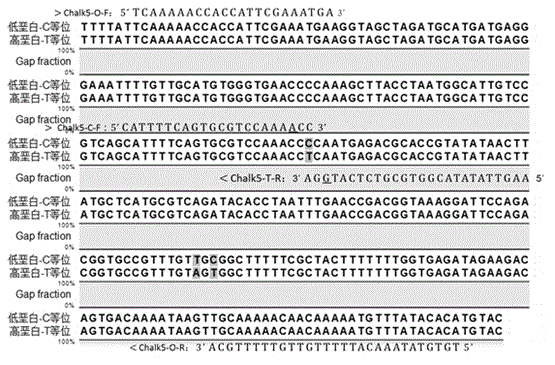
[0036] Aiming at the promoter SNP site "-576-T / C" of the chalky gene Chalk5 (T-high chalky varieties, C-low chalky varieties), use the online tetra-primer ARMS-PCR primer design tool Primer1 (http: / / primer1.soton.ac.uk / primer1.html) Design a functional marker Chalk5-T / C that covers the different sites, and at the same time introduce a mismatch base at the 3'end of the allele-specific primer, Increase specificity ( figure 1 ). The Chalk5-T / C is composed of 4 primers Chalk5-O-F, Chalk5-O-R, Chalk5-C-F and Chalk5-T-R. The primer sequences (5’-3’) are as follows:
[0037] Chalk5-O-F: TCAAAAACCACCATTCGAAATGA
[0038] Chalk5-O-R: TGTGTATAAACATTTTTGTTGTTTTTGCA
[0039] Chalk5-C-F: CATTTTCAGTGCGTCCAAA A CC
[0040] Chalk5-T-R: AAGTTATATACGGTGCGTCTCAT G GA
[0041] (The underlined bases in the primer sequence are the introduced ...
Embodiment 2
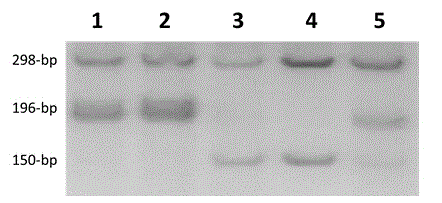
[0044] Example 2 Using the functional marker Chalk5-T / C to analyze the chalky genotypes of 5 rice varieties
[0045] 1. Extraction of rice genomic DNA
[0046] Five rice varieties are used as materials: Minghui 63, Basmati 370, Zhenshan 97B, IR64, Zhenshan 97B / Minghui 63. Select the tender leaves of a single rice plant, and extract rice genomic DNA using the CTAB method. The specific steps are as follows: (1) Take an appropriate amount of fresh leaves and place them in a 2 mL centrifuge tube, add liquid nitrogen to mash, and add 1 mL of CTAB extract to the centrifuge tube. , Shake well; (2) Place in a 65℃ water bath or thermostat, shake gently every 10 minutes, take out after 30~45 minutes; (3) After cooling for 2 minutes, add chloroform-isoamyl alcohol (24:1) (4) Put the centrifuge tube in a centrifuge at 15,000 r / min and centrifuge for 10 minutes and then take it out; (5) Carefully transfer the supernatant to a new sterile centrifuge tube Then add 600μL of pre-cooled i...
Embodiment 3
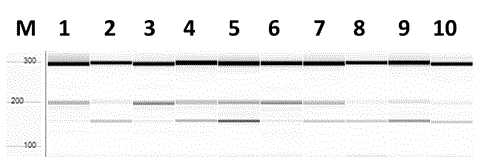
[0065] Example 3 Using Rice Chalkiness Gene Molecular Marker Chalk5-T / C to Identify Low Chalkiness Genotype Rice Single Plant
[0066] 1. Experimental materials: 10 individual rice plants of the F2 offspring of the hybrid rice variety Huazhan and the polymerization line "H463".
[0067] 2. Extraction of Rice Genomic DNA: Same as Example 2.
[0068] 3. Functional marker Chalk5-T / C amplification: the same as in Example 2.
[0069] 4. Capillary electrophoresis detection and genotype determination of amplified products
[0070] Dilute the amplified product and transfer it to Fragment Analyzer® Automated CE System (AATI, USA) for detection, follow the operating instructions of the kit DNF-910-K2000, and use PROSize® 2.0 data analysis software for data analysis, and finally Genotype is judged:
[0071] The amplified products are two fragments of 298 bp and 196 bp, showing that the genotype of the marker individual is C / C homozygous ( image 3 The first, third, and si...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com