Construction method of recombinant Bacillus subtilis expressing d-psicose 3-epimerase based on d-alanine-deficient selection marker
A technology of Bacillus subtilis and epimerase, applied in the field of enzyme genetic engineering, can solve the problems of difficult separation and purification, complicated process, many by-products, etc.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
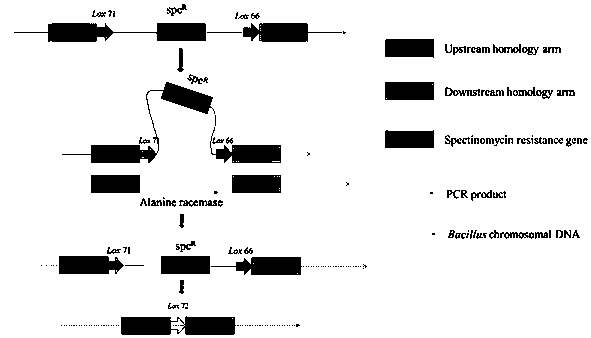
[0023] Example 1: D-alanine-deficient Bacillus subtilis 1A751 ( dal - ) Build
[0024]
[0025] Using plasmid p7S6 as a template, the primer pair P3 / P4 will lox 71- spc - lox 66 Antibiotic resistance gene fragments were amplified and recovered. D-alanine racemase gene on chromosome 1A751 of Bacillus subtilis dal The 800-900bp length fragments on both sides are selected as the homology regions, and the homology regions on both ends are amplified and recovered with primer pairs P1 / P2 and P5 / P6 respectively.
[0026] Use primer pair P1 / P6 to combine the homology arm fragments at both ends with antibiotics lox 71- spc - lox 66 fused together by PCR technology.
[0027] PCR reaction system: Add the following reagents in order to 0.2mL PCR tube: 1.5μL each of the upstream and downstream primers; 5μLhusion HF buffer (5×); 2μL 10mM dNTP mix (2.5mM each); 2μL upstream homology arm; 0.5μL lox 71- spc - lox 66; 2μL downstream homology arm; 0.5μL Phusion high-fidelity DNA polymerase; add d...
Embodiment 2
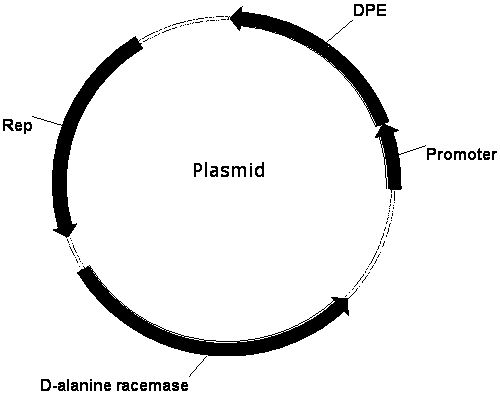
[0032] Example 2: Construction of food-grade safe plasmid pUB-P43-DPE-dal
[0033]
[0034] To enhance expression, a strong promoter P43 is fused upstream of the DPE enzyme gene to form a P43-DPE expression unit. The P43-DPE fragment was amplified using primers P7 / P9. Using pUB110 as the starting vector, using the primer pair P8 / P10, the vector backbone pUB was amplified. Then the P43-DPE and pUB fragments were subjected to PCR to form multimers.
[0035] PCR reaction system: Add the following reagents in order to 0.2mL PCR tube: 5μL Phusion HF buffer (5×); 2μL 10mM dNTP mix (2.5mM each); 2μL P43-DPE; 2μL pUB; 0.5μL Phusion high-fidelity DNA polymerization Enzyme; add distilled water to a final volume of 50μL.
[0036] PCR amplification conditions: pre-denaturation at 98°C for 30s; denaturation at 98°C for 30s, annealing at 55°C for 15s, extension at 72°C for 1 min (30 cycles); extension at 72°C for 10 min.
[0037] The PCR product was directly transformed into host 1A751 and scree...
Embodiment 3
[0039] Example 3: Fermentation of recombinant bacteria
[0040] 1. DPE enzyme activity determination method: In a 1mL reaction system, add 800μL of 100g / L D-fructose dissolved in phosphate buffer (50mM, pH7.0), 200μL of fermentation broth, incubate at 55°C for 10min, then Boil for 10 minutes to stop the enzyme reaction.
[0041] 2. Detect the amount of D-psicose produced by HPLC and calculate the enzyme activity. Enzyme activity unit (U): The amount of enzyme required to catalyze the production of 1μmol D-psicose per minute.
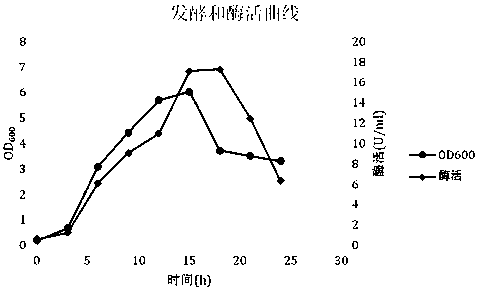
[0042] 3. Use an inoculating loop to pick fresh colonies from the plate and inoculate them into the seed culture medium at 37°C, 200 rpm, and incubate for 12 to 14 hours. Inoculate the fermentation medium with an inoculum of 3%, 37°C, 200 rpm, and sample regularly to determine OD 600 , Enzyme activity data ( image 3 ).
[0043] Seed medium: tryptone (10g / L), yeast extract (5g / L), NaCl (10g / L), prepared with purified water.
[0044] Fermentation medium: glucose...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com