Strong-winter winter rape (Brassica campestris) SOD enzyme molecular marker and QTL locus
A Chinese cabbage-type winter rapeseed and molecular marker technology, applied in the fields of molecular biology and genetic breeding, can solve the problem that the research on molecular markers and QTL mapping has not yet been reported, and achieve the advantages of speeding up the breeding process, saving production costs and improving screening efficiency. Effect
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
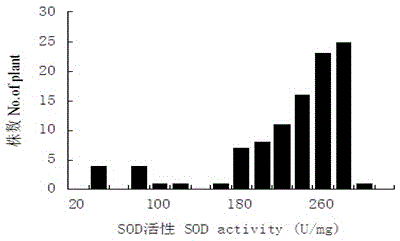
[0032] Example 1: Construction of isolated populations of strong winter Chinese cabbage-type winter rapeseed and determination of SOD enzyme activity.
[0033] The specific construction of the isolated population used in this embodiment is as follows:
[0034] (1) In August 2010, multi-generation bagging and self-crossing of the parent rapeseed 'Longyou 7' with strong winter resistance and strong winter resistance and rapeseed 'Longyou 9' with strong winter resistance were used to configure the hybrid combination, harvested in May 2011 F1 generation seeds.
[0035] (2) F1 seeds were sown in August 2011, and bagged and self-crossed in April 2012 to obtain F2 generation seeds.
[0036] The F2 generation seeds were sown in August 2012. At the seedling stage, 103 plants were randomly selected to be listed and marked, and fresh young leaves were collected in early November, brought back to the laboratory in an ice box and stored in a -70°C ultra-low temperature refrigerator. SOD...
Embodiment 2
[0037] Example 2: Extraction of total DNA from leaves of parents and F2 generation segregation populations.
[0038] Total DNA was extracted from the leaves by the CTAB method, and the specific steps were as follows:
[0039] (1) Pick healthy young leaves as materials for extracting rapeseed genomic DNA, wash them with distilled water, and dry them with paper.
[0040] (2) Take about 0.5g of fresh leaves and put them in a mortar, add liquid nitrogen, grind them quickly until they are whitish powdery, and put them into a 2ml centrifuge tube immediately.
[0041] (3) Add 700ul of preheated 2×CTAB extraction buffer to infiltrate the powder and invert the centrifuge tube to fully disperse the powder. Place in a water bath at 65°C for 60 minutes, during which time gently mix 2-3 times.
[0042] (4) Take out the centrifuge tube, cool to room temperature, add 700ul of chloroform / isoamyl alcohol (24 / 1), gently invert to mix, and let stand for 10min.
[0043] (5) Centrifuge at 13000r...
Embodiment 3
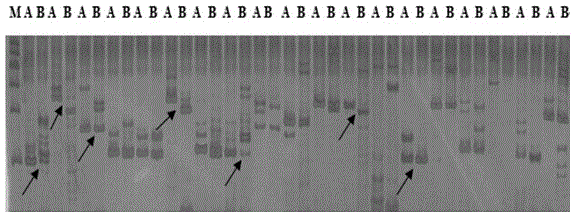
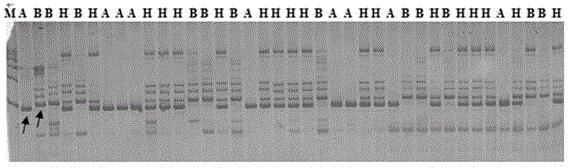
[0051] Example 3: Source, synthesis and polymorphism screening of SSR and InDel primers.
[0052] (1) 315 pairs were uniformly selected from the rapeseed microsatellite primer sequences published on www.UKcrop.net and all primers on 10 chromosomes published on BrassicaDatabase website http: / / brassicadb.org / . Among them, there are 51 pairs of SSR primers and 264 pairs of InDel primers. Primers were synthesized by Shanghai Bioengineering Technology Co., Ltd.
[0053] (2) Randomly select 6 strains of DNA from each parent and mix them in equal amounts, and use them as templates for screening primers.
[0054] (3) PCR reaction system
[0055] The PCR reaction system is 10ul:
[0056] MixTaq enzyme 6ul
[0057] Upper primer 0.5ul
[0058] Lower primer 0.5ul
[0059] ddH2O2ul
[0060] Template: 1ul
[0061] (4) PCR amplification program: pre-denaturation at 94°C for 4 minutes; denaturation at 94°C for 50s, annealing at 55-60°C for 50s (annealing temperature decreased or incre...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com