Indel mark of powdery mildew resistance allele er1-7 of peas and application thereof
A powdery mildew resistance gene, pea technology, applied in the field of plant pathology and crop resistance genetic breeding, can solve the problem that the effectiveness of functional markers has not been verified
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0055] Example 1, the acquisition of the Indel marker InDel111-120 of the present invention
[0056] 1.1er1-7 feature flag development
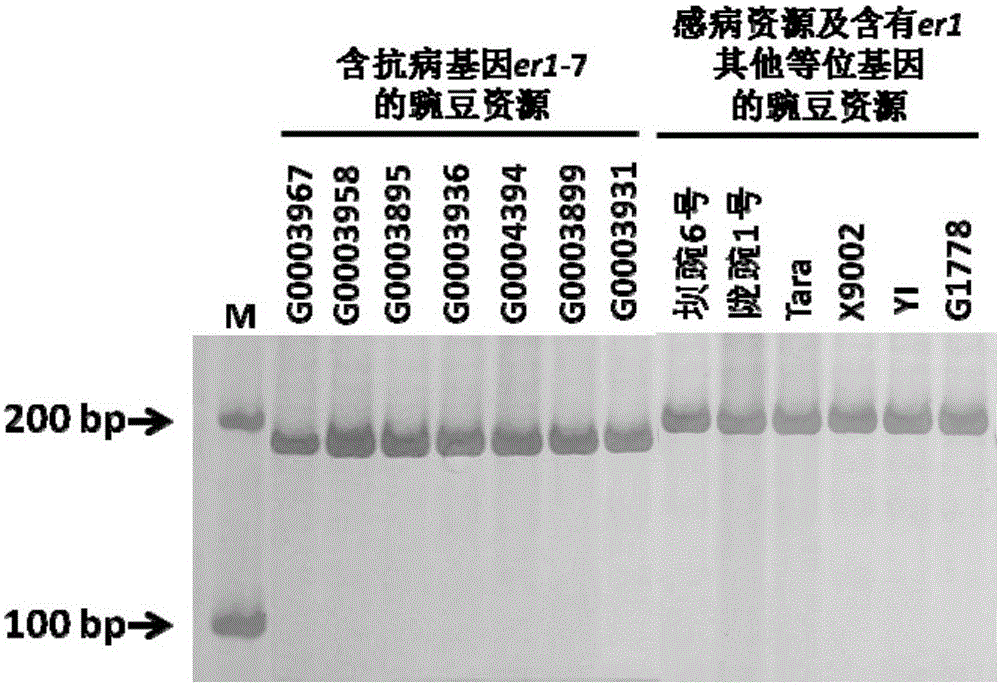
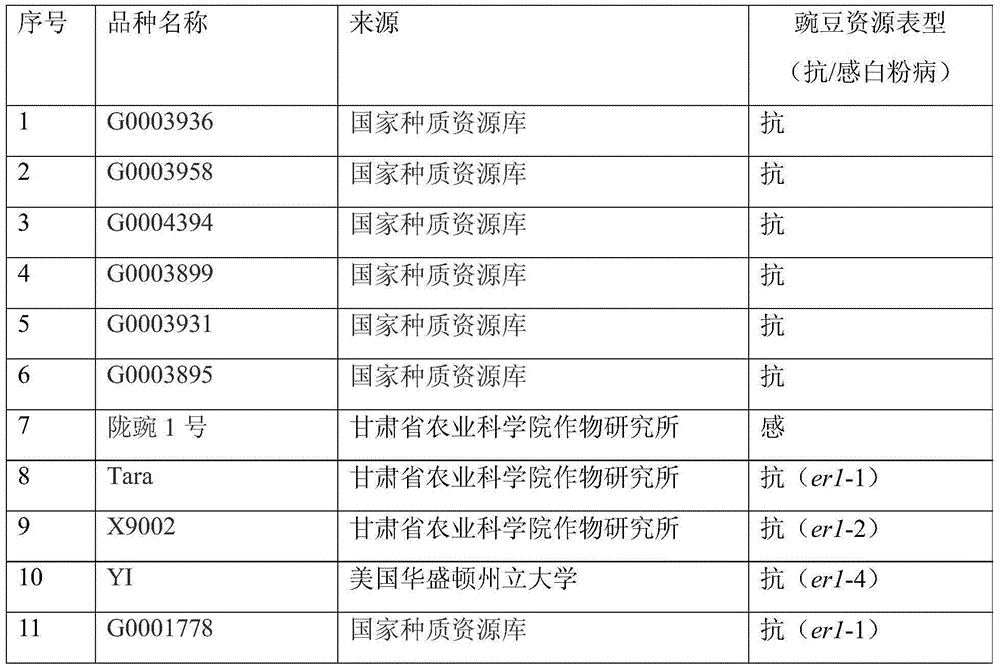
[0057] According to the comparative analysis of the PsMLO1 candidate gene sequences of the resistant variety G0003967 and the susceptible variety Bawan 6 and the wild-type PsMLO1 sequence (FJ463618) (Humphry et al., 2011), it was found that the open reading frame 111-120 of the PsMLO1 cDNA sequence of G0003967 The base deletion, which has not been reported yet, is a new variant of PsMLO1. This result indicates that G0003967 contains a new powdery mildew resistance allele, named er1-7. According to the 10bp deletion on the first exon of the PsMLO1 gene of G0003967, the upstream and downstream primers were designed with PrimerPremier5.0 software to develop InDel functional markers, and the Indel markers were named InDel111-120.
[0058] The nucleotide sequence of the upstream and downstream primers of the Indel marker InDel111-120 is as follow...
Embodiment 2
[0070]Embodiment 2, the assembly of kit of the present invention
[0071] Based on the Indel marker InDel111-120 obtained in Example 1, the kit described in the present invention was assembled. The kit includes upstream and downstream primers having sequences shown in SeqID No.1 and SeqID No.2 for amplifying the Indel marker InDel111-120.
[0072] The kit also includes conventional reagents for PCR reactions and / or electrophoresis. Specifically, the conventional reagents used for PCR reaction include: dNTPs, TaqDNA polymerase, Mg 2+ , PCR reaction buffer, standard positive template, double distilled water, etc., can also be common commercially available PCR kits, PCR reaction Mix, etc.; the conventional reagents for electrophoresis include: TE buffer, polyacrylamide , agarose, tetramethylethylenediamine, formaldehyde, silver nitrate, etc.
Embodiment 3
[0073] Example 3, Verification and application of the Indel marker or kit of the present invention
[0074] 1.1 Validity verification and screening application of the Indel marker or kit of the present invention in screening hybrid offspring containing gene er1-7
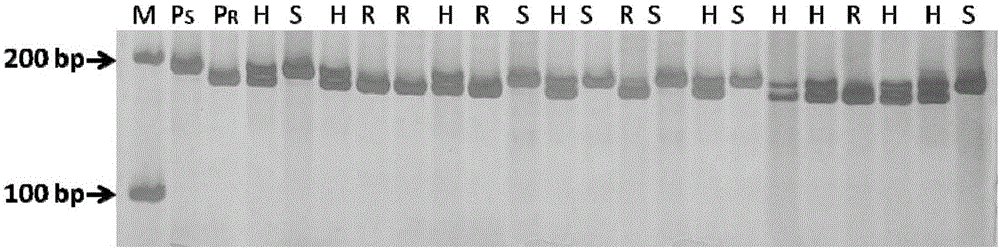
[0075] Use the Indel marker InDel111-120 obtained in Example 1 or the kit obtained in Example 2 for population verification to identify the F 1、 f 2 The genotype of each individual plant in the population. InDel111-120 in all F 1 Two target bands were amplified in the plants, which were 183bp and 193bp respectively. InDel111-120 in F 2 There are three situations in the amplification results of individual plant materials in the population: homozygous disease-resistant genotype (183bp), homozygous disease-resistant genotype (193bp) and heterozygous genotype (183bp, 193bp). Band type and F 2 There is a one-to-one correspondence between the traits and phenotypes of the population. The three genotypes shown corres...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com