Kit and primer pair group for detecting expression of cytosine deaminase and molecular genes related to cytosine deaminase in free DNA (deoxyribonucleic acid) in peripheral blood
A cytosine deaminase and gene expression technology, which is applied in the field of a kit for detecting the gene expression of cytosine deaminase and its related molecules in peripheral blood free DNA and a primer pair set, which can solve the heterogeneity of tumor cells , can not effectively repair mutations, low activity and other problems, to achieve the effect of improving sensitivity
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0064] Embodiment 1: Establish detection method
[0065] 1) Sample preparation:
[0066] Extract peripheral blood and extract cell-free DNA;
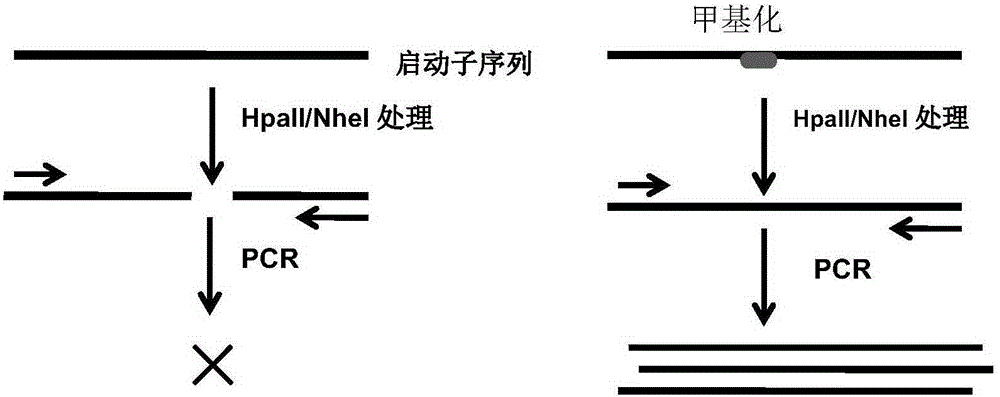
[0067] Use HpaII endonuclease to digest free peripheral blood DNA. The enzyme digestion system is shown in Table 1; the enzyme digestion condition is: 37°C for 2 hours; after HpaII enzyme digestion, further inactivate the enzyme digestion reaction system , the inactivation condition is: inactivation at 65°C for 20min. The inactivated products are used as templates for real-time fluorescent quantitative PCR (qPCR) amplification reactions of Apobec3B / C, Apobec3F / G and UNG genes.
[0068] Use NheI endonuclease to digest free peripheral blood DNA. The enzyme digestion system is shown in Table 2; the enzyme digestion conditions are: 37°C for 2 hours; after NheI enzyme digestion, further inactivate the enzyme digestion reaction system, the inactivation conditions For: 65 ℃ inactivation 20min. The inactivated product is used as a template ...
Embodiment 2
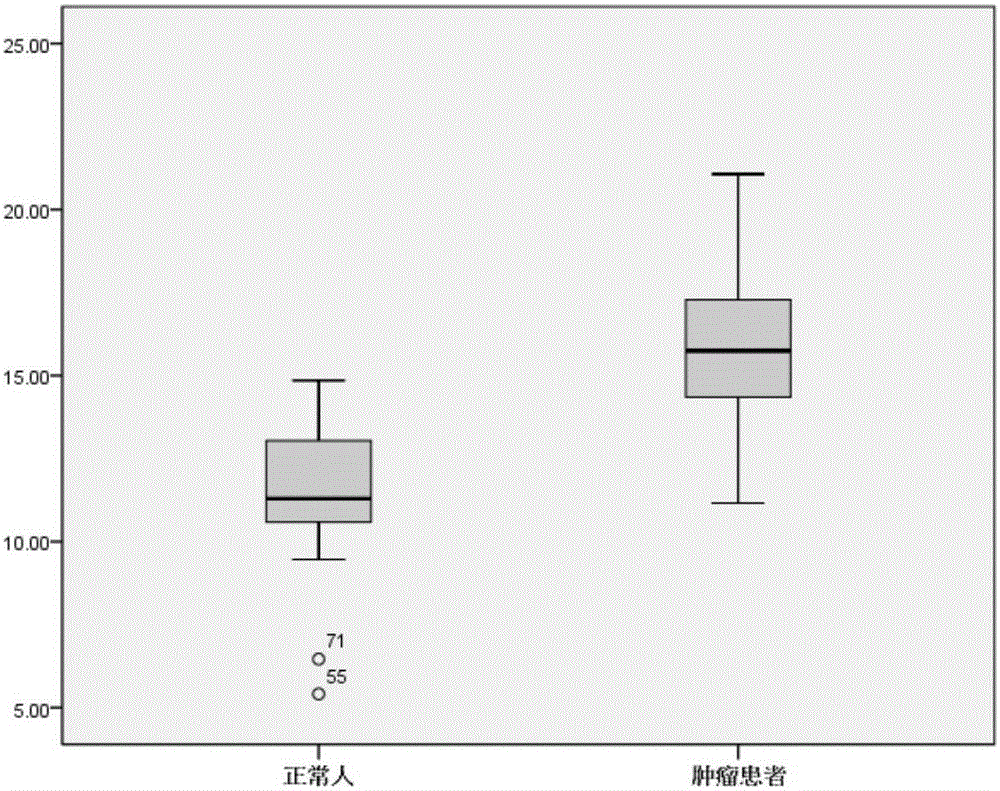
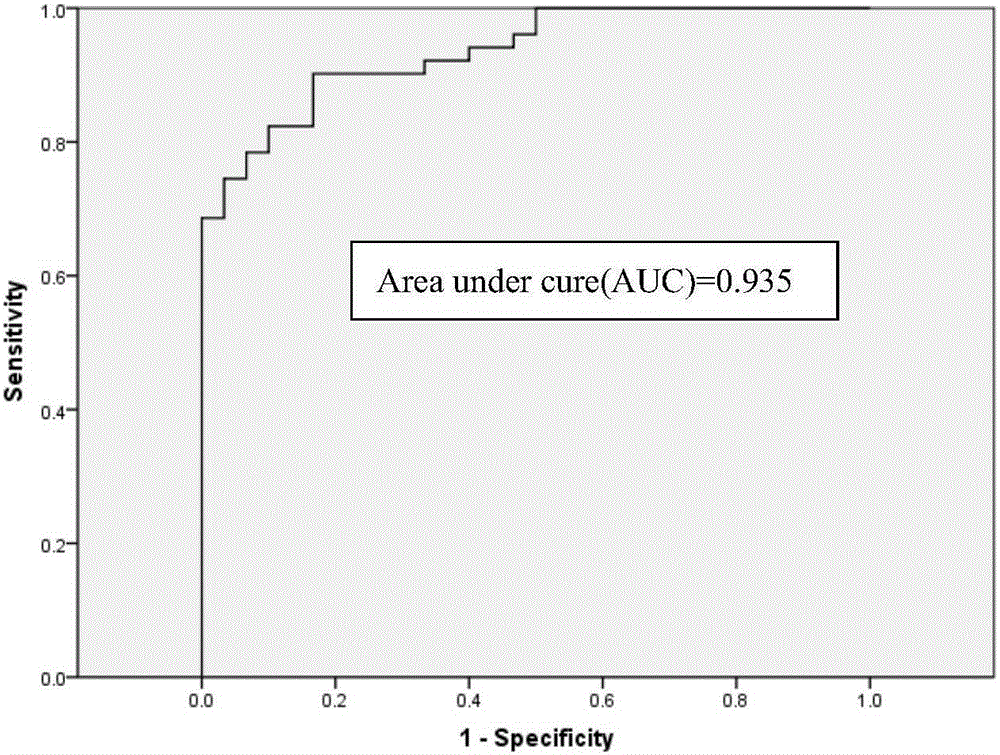
[0124] According to the method of implementation 1, the Apobec3B / C, Apobec3F / G, UNG and AID gene mutations in normal people and tumor patients were detected, and the ΔΔCt weighted value was calculated. The △△Ct weighted value of cancer patients is shown in Table 12, and the △△Ct weighted value of cancer patients is shown in Table 13.
[0125] Table 12: Control group (30 cases):
[0126]
[0127]
[0128] Table 13: Patients in Tumor Group
[0129]
[0130]
[0131] There were 11 cases of lung cancer, 10 cases of liver cancer, 9 cases of gastric cancer, 5 cases of colorectal cancer, 3 cases of esophageal cancer, 2 cases of pancreatic cancer, 1 case of breast cancer, and 10 cases of other tumors.
[0132] According to statistical analysis, the CutOff value is set to 13, so that 6 of the 30 normal subjects (samples 8, 12, 14, 17, 20 and 23) have a slightly higher △△Ct weighted value than 13; In 4 cases (samples 8, 9, 38 and 40), the weighted value of ΔΔCt was lower t...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com