Method for rapidly identifying species of salted rhopilema esculentum kishinouye
A pickling and fast technology, applied in the direction of biochemical equipment and methods, microbial determination/inspection, etc., to achieve the effect of high accuracy, strong specificity and high sensitivity
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0053] 1. Desalinize the pickled cotton stings, change the water once every 12 hours, and change 4 times in total, use ethanol solution as the stock solution, store the dehydrated cotton stings in a solution with a final concentration of 70% ethanol (v / v), and store ;
[0054] 2. Take out 150 mg of the jellyfish stings stored in the ethanol solution, cut them into pieces, centrifuge at 10,000 rpm for 5 minutes, put them on a dust-free paper to absorb the water, and use the deep-processed food DNA extraction kit to extract jellyfish genomic DNA;
[0055] 3. Use 16SrRNA gene universal primers for PCR amplification, and store the product at -20°C;
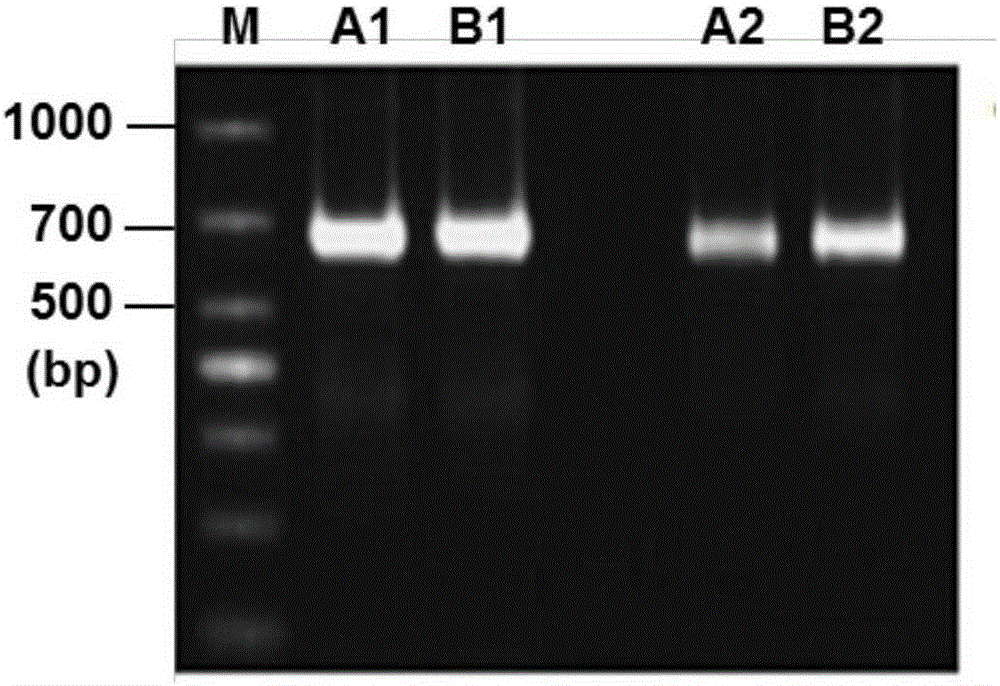
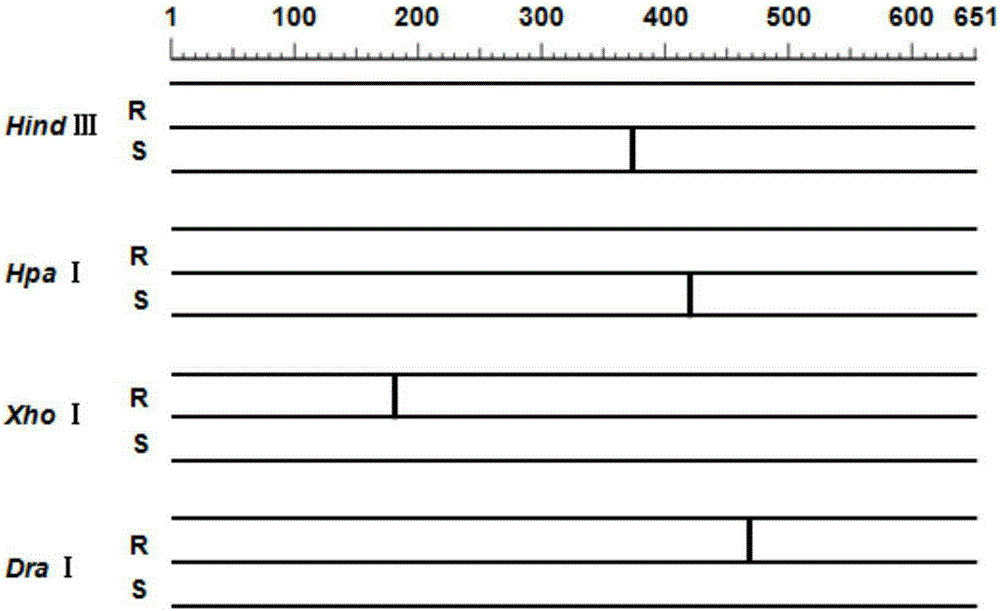
[0056] 4. The PCR amplification product was digested with HindⅢ, HpaI, DraI and XhoI endonucleases, and the digested products were separated by 3% agarose gel electrophoresis and detected by imaging.
[0057] The result is as Figure 5 Shown, wherein, A represents HindⅢ, B represents HpaI, C represents XhoI, D represents DraI. It c...
Embodiment 2
[0060] 1. Purchase 4 kinds of commercially available pickled jellyfish from Shandong and Liaoning, numbered 1, 2, 3, 4, respectively, and carry out desalination treatment, change the water once every 12 hours, and change the water 4 times in total. Use ethanol solution as the stock solution, and dehydrate the jellyfish. The jellyfish is stored in a solution with a final concentration of 70% ethanol (v / v), for future use;
[0061] Among them, 1 represents the pickled jellyfish purchased from Yantai, Shandong, 2 represents the pickled jellyfish purchased from Yingkou, Liaoning, and 3 and 4 both represent the pickled jellyfish purchased from Dalian, Liaoning.
[0062] 2. Take out 150 mg of jellyfish stored in ethanol solution, cut it into pieces, centrifuge at 7500rpm for 10 minutes, put it on a dust-free paper to absorb the water, and use the deep-processed food DNA extraction kit to extract jellyfish genomic DNA;
[0063] 3. Use 16SrRNA gene universal primers for PCR amplificat...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com