Method for high specificity detection of target DNA in sample, and application thereof
A technology for detecting samples and high specificity, applied in the field of nucleic acid detection, can solve the problems of false positive detection results, high hybridization background, restricting the wide application of non-isotope labeled probe methods, etc., to improve specificity, improve detection efficiency, avoid The effect of false positives
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0086] (1) Immobilization of target DNA on the surface of solid support
[0087] 1.1 Experimental materials:
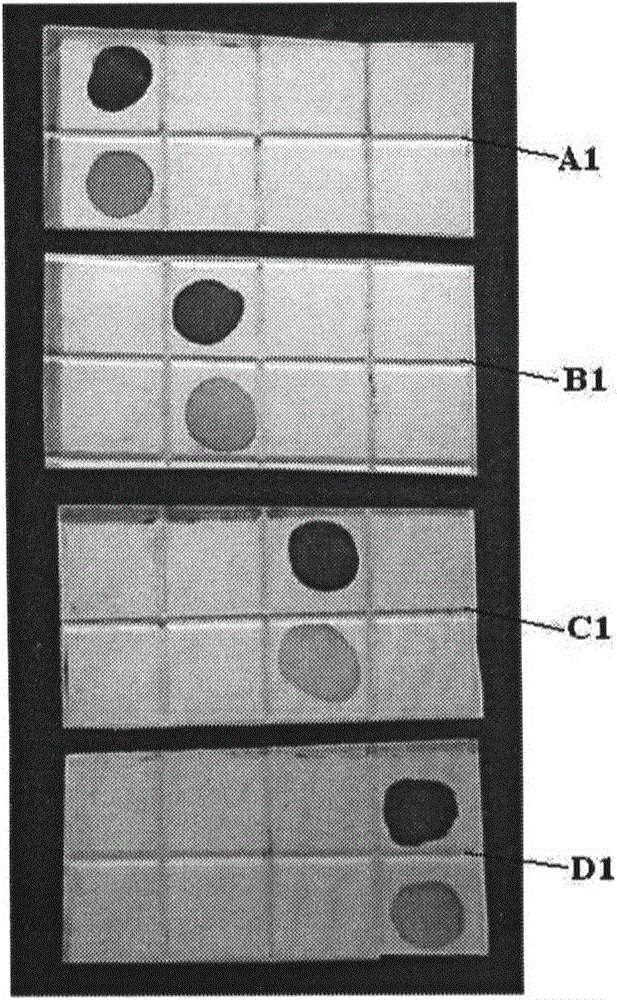
[0088] Solid support: nitrocellulose membrane, produced by Millipore, 0.45μm pore size, cut into 4 membranes with a size of 2cm×1cm for use, and 4 nitrocellulose membranes are labeled A1, B1, and C1 And D1.
[0089] Test sample containing target DNA: human papillomavirus (HPV) type 16 (1pg / μl and 0.1pg / μl), type 18 (1pg / μl and 0.1pg / μl), type 6 (1pg / μl and 0.1pg / μl) and type 11 (1pg / μl and 0.1pg / μl) genome-wide plasmid standards, provided by Shanghai Jereh Company.
[0090] Denaturing liquid: 0.4mol / L NaOH solution.
[0091] 20×SSC buffer solution: 3.0mol / L NaCl, 0.3mol / L sodium citrate, pH 7.0.
[0092] In addition, the 20×SSC buffer solution was diluted to 15×SSC buffer solution and 10×SSC buffer solution for use.
[0093] 1.2 Experimental steps
[0094] 1.2.1 Pretreatment of nitrocellulose membrane: Use tweezers to take the nitrocellulose membrane and soak it in 15×SSC buffe...
Embodiment 2
[0153] Same as Example 1, except that the composition of the pretreatment solution in Example 1 is adjusted to: PH7.0, 0.1mol / LTris-HCl, 1mol / L NaCl, 3% BSA, 0.05% TritonX-100, 0.1% SLS and 0.1% APAM, the balance is water. Other conditions remain unchanged.
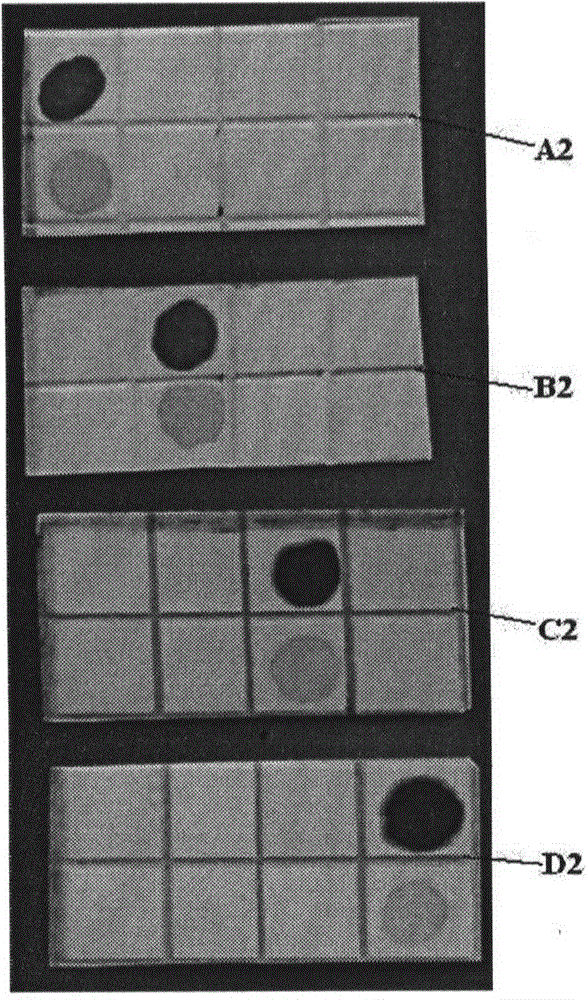
[0154] In this example, the 4 nitrocellulose membranes are marked as A2, B2, C2, and D2, and the color results are as follows figure 2 Shown.
Embodiment 3
[0156] Same as Example 1, except that the composition of the pretreatment solution in Example 1 is adjusted to: PH8.0, 0.1mol / LTris-HCl, 1mol / L NaCl, 4% BSA, 0.2% Tween-20, 0.2% SLS, 0.3% APAM, the balance is water. Other conditions remain unchanged.
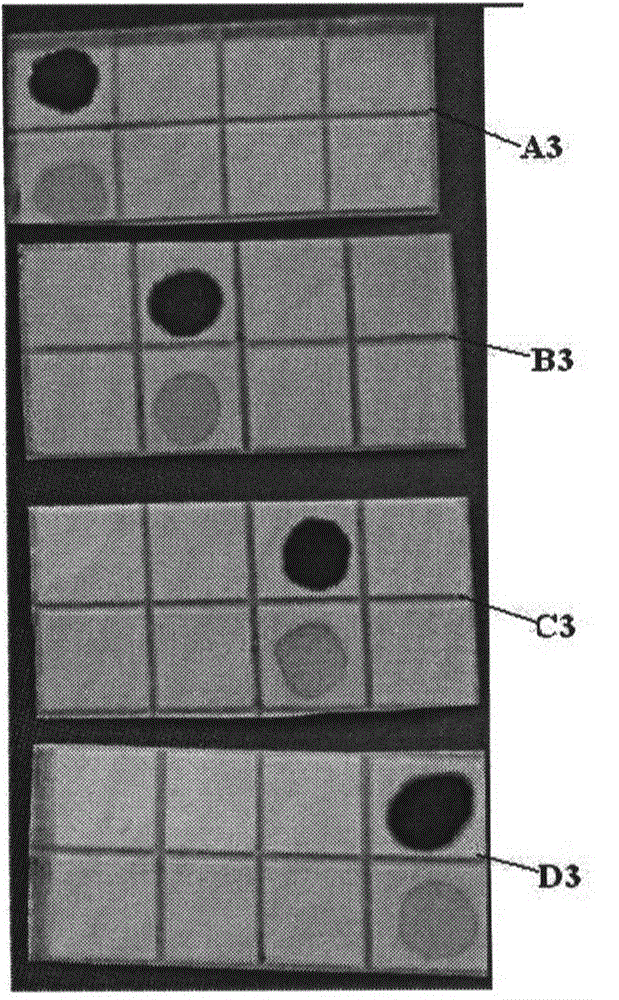
[0157] In this example, the 4 nitrocellulose membranes are marked as A3, B3, C3 and D3, and the color rendering results are as follows image 3 Shown.
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com