Rapid and sensitive genotype identification and nucleic acid detection
A nucleic acid molecule and oligonucleotide probe technology, which is used in the fields of rapid and sensitive genotype identification and nucleic acid detection, and can solve the problems of limited membrane area and long incubation time.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
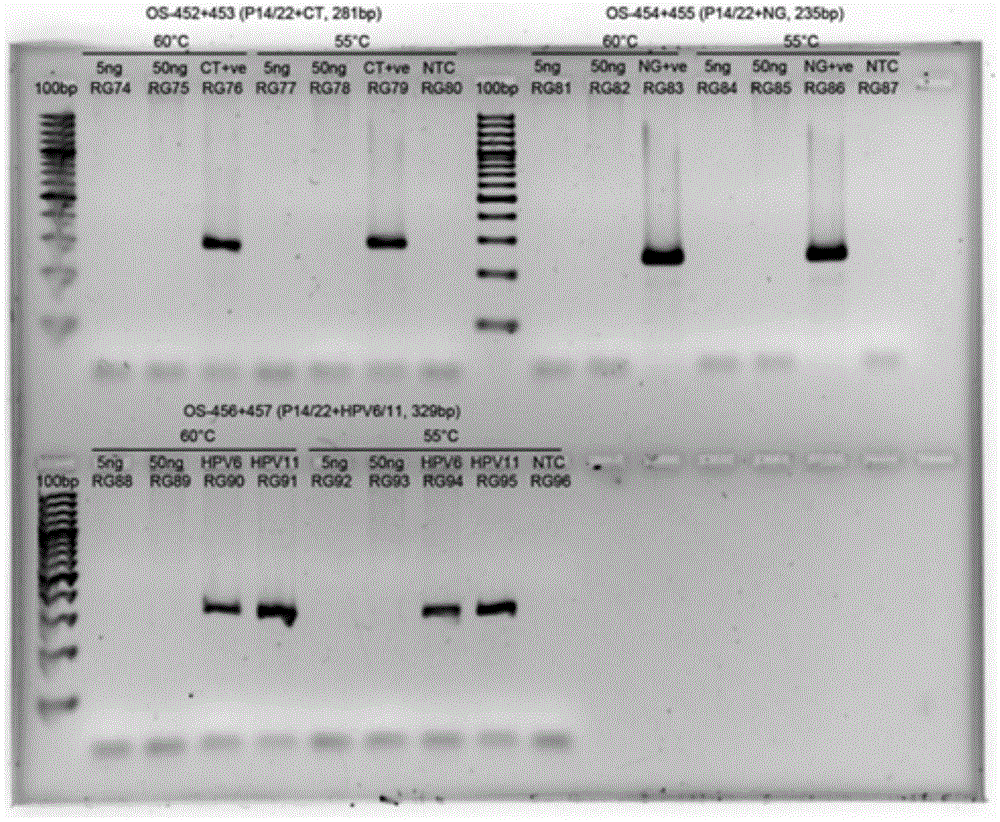
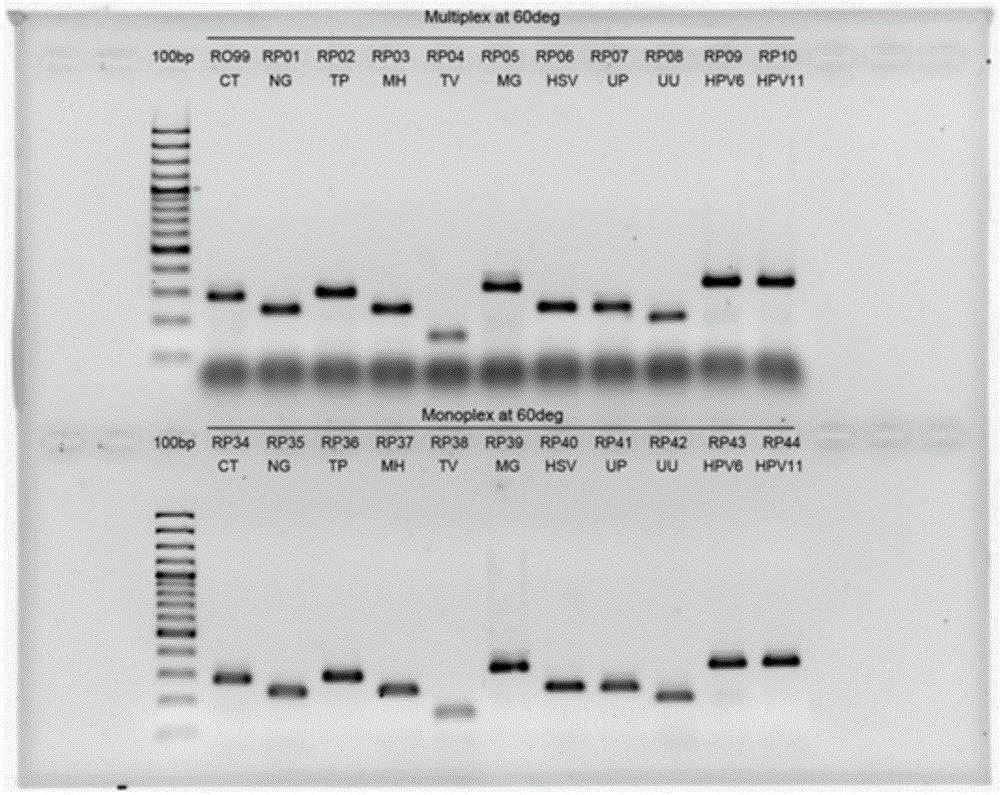
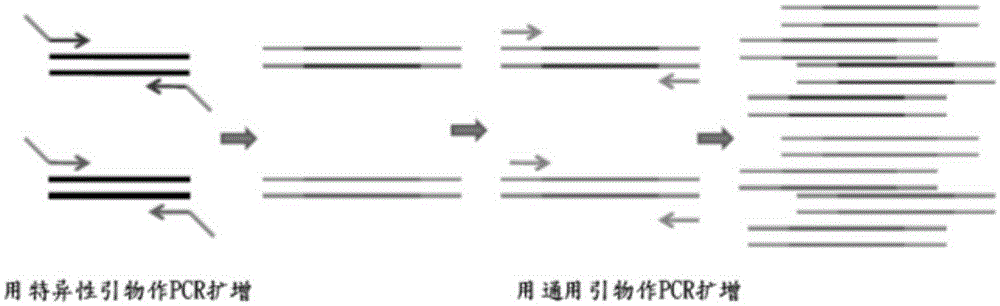
[0074] test program
[0075] The separation steps for DNA, PCR amplification, hybridization, detection samples and result analysis, such as WO / 2011 / 139750 or related procedures and techniques known to those skilled in the art can be applied to the present invention. Various internal controls (IC) can be included in the test test to verify the DNA sample, PCR amplification and color development of the results. The size and format of each guide film array can be used according to individual needs, or used to optimize the use of the reaction chamber to maximize cost savings.
Embodiment 2
[0077] Genotyping MDR-TB
[0078] In one embodiment, the present invention provides methods and kits for DR-MTB genotyping. The method and kit use polymerase chain reaction (PCR) and "drainage" hybridization technology to detect Mycobacterium tuberculosis (MTB) resistant to rifampicin (RIF) and isoniazid (INH) . The corresponding primers used to amplify the rpoB gene, katG gene and INHA gene have been confirmed not to cross-react with the human genome. RS-IAC is an internal control primer that is not targeted at the human or MTB genome and is used to monitor the PCR amplification process. The gene mutations used to detect drug resistance include rpoB gene (D516V, D516G, H526D, H526Y, H526L1, S531L and S531W), katG gene (S315T1 and S315T2), and inhA gene (-15TC / T). There are also five probes for detecting wild-type MTB mutations, which helps to verify whether the PCR amplification reaction of rpoB gene, katG gene and inhA gene has been completed. In one embodiment, the primer...
Embodiment 3
[0097] Beta Mediterranean Genotyping
[0098] The present invention provides a systematic detection of β-globulin gene mutations. The gene mutations include TATA-28 (A> G), TATA-29 (A> G), start codon (G> A), detection of codon 5 (-CT), codon 8 / 9 (+G), codon 15 (G> A), codon 16 (-C), codon 17 (A> T), codon 19 (A> G), codon 26 (G> A) (hemoglobin E), codon 27 / 28 (+ three), codon 30G> C, IVS1.1 (G> T), IVS1.1 (G> A), IVS1.5 (G> C), codon 41 / 42 (-TCTT), codon 43 (G> T), codon 71 / 72 (+A), IVS2.1 (G> A), IVS2.654(C> T) and a deletion of 619 bp.
[0099] The present invention also provides a method for detecting β-globulin mutations in a sample of a subject, the method comprising the following steps: (a) obtaining a sample containing template nucleic acid molecules from the subject; (b) with a sequence number selected from : The set of 201-205 primers amplifies the template nucleic acid molecule, thereby generating an amplified product; and (c) the amplified product is composed of an olig...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com