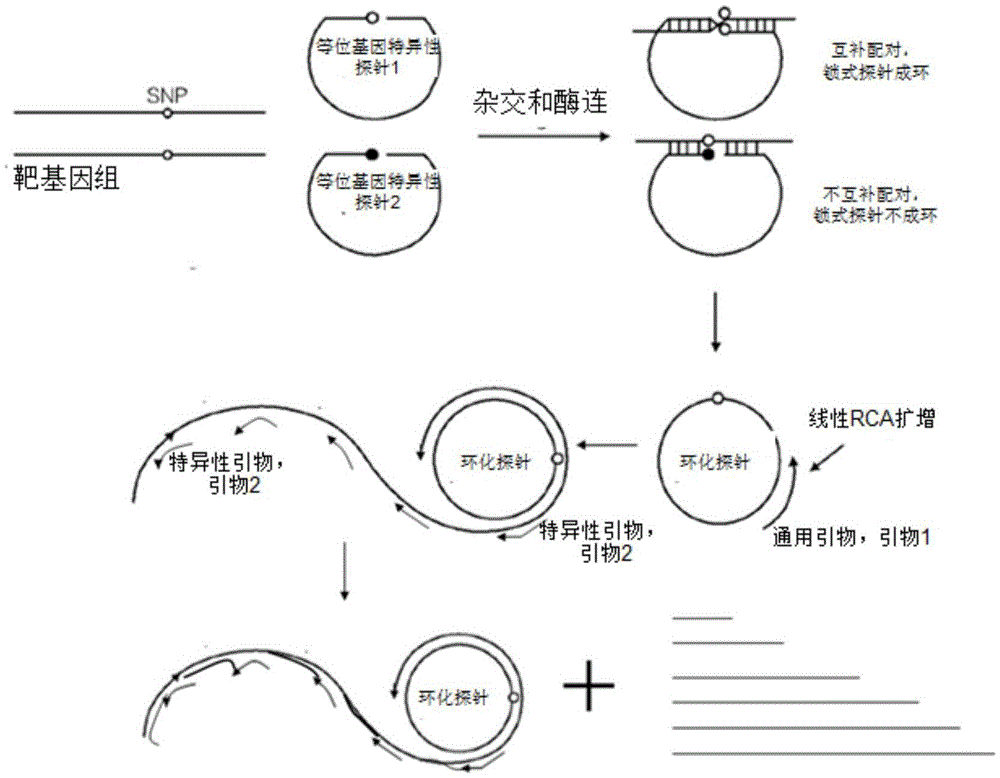
SNP (single nucleotide polymorphism) typing detecting method for utilizing locking-type probe for rolling ring amplification
A padlock probe and rolling circle amplification technology, applied in the biological field, can solve the problems of increasing the probability of operator infection, high cost, cumbersome operation, etc., shorten the operation steps and detection time, reduce the risk of cross-infection, and detect accurate results
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0026] 1. Design of padlock probes and amplification primers
[0027] Obtain all reference genotypes of the SNP site (rs27044) on the ERAP1 gene from the genebank database, and obtain the gene sequence of 500 bp upstream and downstream of the single nucleotide polymorphism site, and use the primer5.0 software to design two allele-specific A sex padlock probe (to simplify the description, it will be referred to simply as a padlock probe hereinafter) and two amplification primers. in:
[0028] Detection of rs27044 site: two padlock probes (as shown in SEQ ID NO: 1, 2): Pro-15'-TGAGGCGTCGTAAGCCATAAGTAGGATAGGACAGGAAAGAAACGCTCCTCATCATCAAG-3', Pro-25'-TGAGGCGTCGTAAGCCATAAGTAGGATAGGACAGGAAAGAAACGCTCCTCATCATCAAC-3'; two amplification primers (such as SEQ ID NO: 3 , 4): the first primer 5'-CTTTTCCTGTCCTATCCTACTTATG-3', the second primer 5'-CTCCTCATCATCAACTG-3';
[0029] 2. Padlock probe phosphorylation
[0030] Phosphorylate the 5′ end of the designed allele-specific padlock probe, ...
Embodiment 2
[0041] In order to verify the detection effect of a SNP typing detection method using padlock probe rolling circle amplification of the present invention, the applicant carried out the operation of Example 2, specifically:
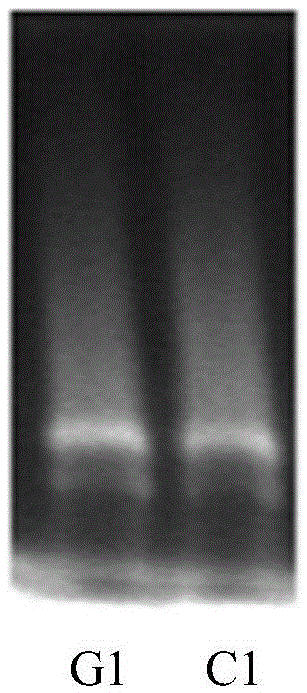
[0042] Take sample 1, purify the DNA genome, and use the SNP typing detection method of the present invention to detect, and the detection results are as follows: figure 2 Shown; And by adopting existing recognized Sanger method to carry out SNP typing detection to peripheral blood sample I to carry out contrast, then test result shows, the detection result of the present invention's detection method is consistent with the detection method of existing recognized Sanger method;
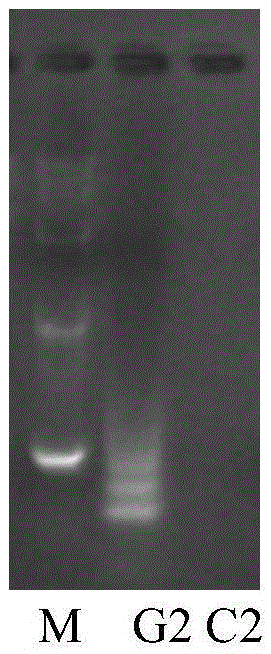
[0043] Take sample II, purify the DNA genome, and use the SNP typing detection method of the present invention to detect, and the detection results are as follows: image 3 shown; and by adopting the existing recognized Sanger method to carry out SNP typing detection on the periph...
Embodiment 3
[0045] In order to verify the detection effect of a SNP typing detection method using padlock probe rolling circle amplification of the present invention, the applicant carried out the operation of Example 2, specifically:
[0046] Get the peripheral blood sample I and use the SNP typing detection method of the present invention to detect, and the detection results are as follows: Figure 4 Shown; The result corresponding to this embodiment is figure 2 ;
[0047] Take peripheral blood sample II and use the SNP typing detection method of the present invention to detect, and the detection results are as follows: Figure 4 Shown;; The result corresponding to this embodiment is Figure 4 ;
[0048] Therefore, the detection result of the detection method of the present invention is accurate and feasible.
[0049] In addition, it should be noted that the percentage of each component in the present invention: the solid-liquid system is the mass percentage, and the liquid-liquid ...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com