Recombinant strain for expressing heterogenous Omega-3 desaturase in mortierella alpina and construction method thereof
A technology of Mortierella alpina and saturase, which is applied in the field of bioengineering and can solve the problems of unreachable EPA production
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0047] Example 1: Codon optimization and artificial synthesis of ω-3 desaturase gene derived from Pythium melon
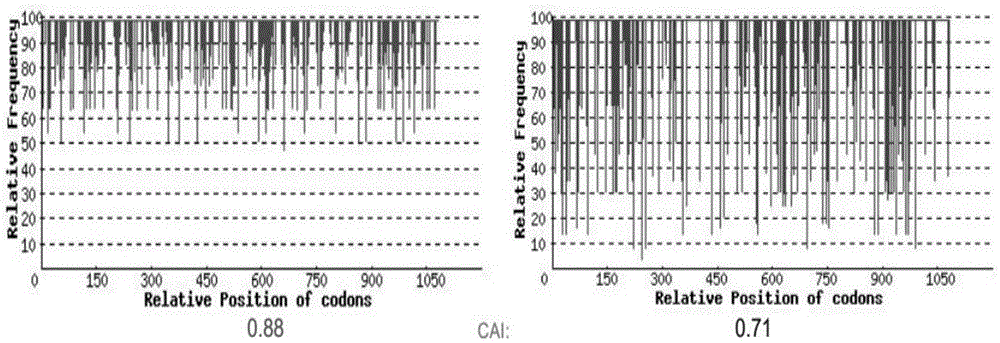
[0048] Since the codon usage preference of the natural PaD17 gene sequence is different from that of the host Mortierella alpina, according to the codon usage preference of Mortierella alpina, the nucleic acid sequence of PaD17 was codon-optimized by GenscriptOptimumGeneTMsystem, and the optimized sequence was synthesized artificially. The latter gene sequence oPaFADS17, as shown in SEQNo.1 and 2, has added HindIII and XhoI restriction sites on both sides, and is connected with the PUC57 carrier to obtain PUC57-oPaFADS17, which is stored in Escherichia coli Top10 (GenScript, Nanjing, China) ,China). The codon availability index and codon usage frequency distribution of genes before and after optimization are as follows: figure 1 shown. In the figure, when CAI is 1.0, it means perfect in the desired expression system, and >0.89 means excellent, with high gene expr...
Embodiment 2
[0049] Example 2: Enzymatic cleavage reaction
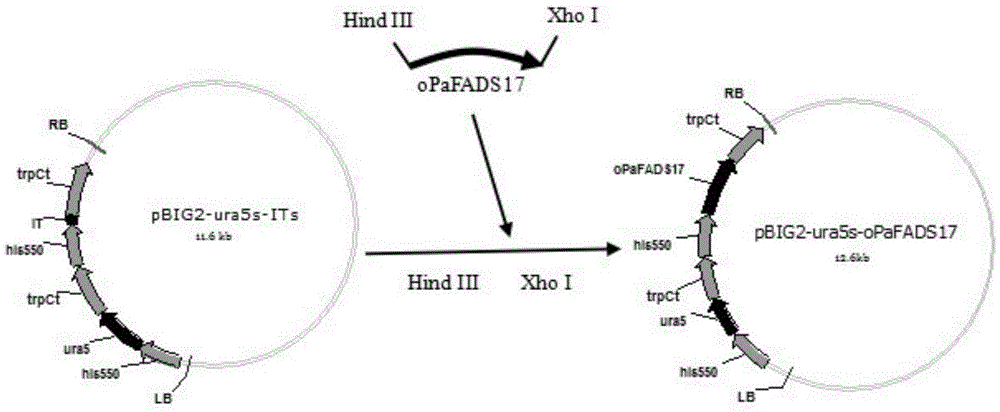
[0050] At 37°C, use the restriction enzyme HindIII to digest the plasmid PUC57-oPaFADS17 and the vector pBIG2-ura5s-ITs fragment overnight with the restriction enzyme HindIII. 58 μL of deionized water, incubate at 37°C for 12h.
[0051]Among them, the vector pBIG2-ura5s-Its is directly obtained according to the content disclosed in Chinese patent application CN201310524221.4. The HPH expression unit was obtained from the pD4 plasmid by PCR, digested with the restriction enzymes EcoRI and XbaI, and inserted into the multiple cloning site (MCS) of pET28a(+) cut by EcoRI and XbaI. , the plasmid pET28a-HPHs was obtained. The ura5 (orotate phosphoribosyltransferase; OPRTase) gene was obtained from the cDNA of Mortierella alpina by PCR, and the ura5 gene was digested with the restriction enzymes BspHI and BamHI, and the digested ura5 gene was inserted into NcoI and BamHI. The plasmid pET28a-HPHs digested with BamHI was replaced with...
Embodiment 3
[0054] Example 3: Ligation reaction
[0055] The purified ω-3 desaturase gene fragment oPaFADS17 was ligated with the vector pBIG2-ura5s-ITs with T4 ligase, and ligated at 4°C for 12 h to obtain the recombinant expression vector pBIG2-ura5s-oPaFADS17. The ligation system was (10 μL): 2 μL of the target gene digested fragment, 3 μL of the vector digested fragment, 1 μL of ligase buffer, 1 μL of T4 ligase, 3 μL of sterile water, and ligated at 4°C for 12 h.
[0056] The ligation product was transformed into E. coli TOP10 competent cells, and the transformation method was as follows:
[0057] (1) Take 100 μL of competent cells under sterile conditions, add 1-2 μL of ligation product, and mix by pipetting.
[0058] (2) Transfer the mixed competent cells into the pre-cooled rotor cup to avoid bubbles.
[0059] (3) Put the electroporation cup into the Bio-Rad electroporator, adjust to the appropriate preset program gear, and electroporate. The voltage condition is 1.8kv.
[0060...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com