Application of SRGN gene in serving as serum marker for detecting triple-negative breast cancer
A triple-negative breast cancer, marker technology, applied in the field of biomedicine, can solve problems that have not been studied and reported
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
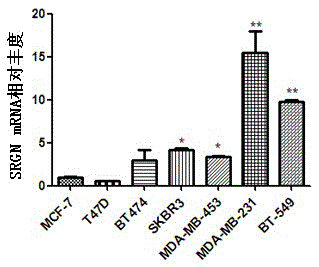
[0039] Example 1 Real-time quantitative PCR analysis of SRGN expression in breast cancer cell lines
[0040] 1. Prepare cDNA
[0041] Take various cultured cells that grow to the logarithmic phase, and extract total RNA by TRIZOL method. After DNaseI digestion, RNA was measured.
[0042] Using oligdT as the primer, the same amount of RNA was reverse transcribed into cDNA according to the Fermentas ReverseTranscriptionSystem program.
[0043] 2. PCR detection
[0044] Using cDNA as a template, follow the SYBGreen operating instructions and use the following primers for real-time quantitative PCR amplification:
[0045] (1) Primer of SRGN gene coding region (amplified fragment is 100bp):
[0046] Forward primer SRGN-F (shown in SEQ ID NO.3):
[0047] 5'-GCTACTCAAATGCAGTCGGC-3';
[0048] Backward primer SRGN-R (shown in SEQ ID NO. 4):
[0049] 5'-CCATTGGTACCTGGCTCTCC-3'.
[0050] (2) The primer amplified fragment of GAPDH gene coding region is 156bp:
[0051] Forward primer GAPDH-F (shown in SE...
Embodiment 2
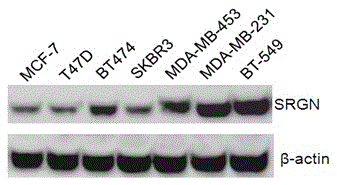
[0058] Example 2 Westenblot analysis of SRGN expression in breast cancer cell lines
[0059] 1. Westenblot
[0060] Take the cultured cells that have grown to the logarithmic phase, use fresh cell lysate containing protease inhibitors to lyse the cells to extract total protein. The protein concentration was determined by the Bradford method of Bio-Rad. Add 40μg protein to each lane to calculate the cell protein volume, add 6×SDS loading buffer at a ratio of 1:5 to dissolve it, denature it at 100°C for 5min, immediately place it on ice for 5min, and centrifuge briefly. After loading the sample, perform electrophoresis. First, electrophoresis at 80V until the bromophenol blue dye enters the separation gel, and then adjust the voltage to 120V to continue electrophoresis until the bromophenol blue reaches the bottom of the gel. The Bio-Rad electrophoresis instrument conducts electrotransfer of protein at 120mV constant pressure for 90min, so that the separated protein is transferred...
Embodiment 3
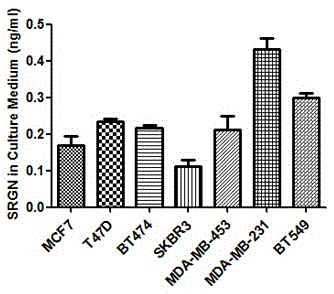
[0063] Example 3 Enzyme-linked immunoassay (ELISA) analysis of SRGN expression in the supernatant of breast cancer cell lines
[0064] 1. Prepare the cell culture supernatant: After digesting the target cells into a single cell suspension and counting, add 1×10 6 Cells / well were seeded in a 6-well plate with a final culture volume of 2ml. After 48 hours of culture, the culture supernatant was collected, and the supernatant was collected after centrifugation at 2000 rpm / min for ELISA experiments.
[0065] 2. Brief steps of ELISA experiment:
[0066] (1) All reagents in the kit are slowly balanced to room temperature before use to avoid rapid dissolution and protein degradation.
[0067] (2) Prepare standard diluent: add 1ml standard diluent to the standard, let it stand at room temperature for 10 minutes, and rub it repeatedly to promote dissolution. At this time, the concentration is 20ng / ml. Then prepare 7 standard dilution EP tubes, add 250μl standard dilution solution to each EP t...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com