DNA probe, reagent kit and method for detecting deoxyribonuclease
A deoxyribonucleic acid and DNA probe technology, applied in the field of enzyme detection, can solve the problems of large influence and difficult detection technology, and achieve the effects of high sensitivity, small reaction volume and low cost
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0042] Embodiment 1: the preparation of probe
[0043] 1 sequence design
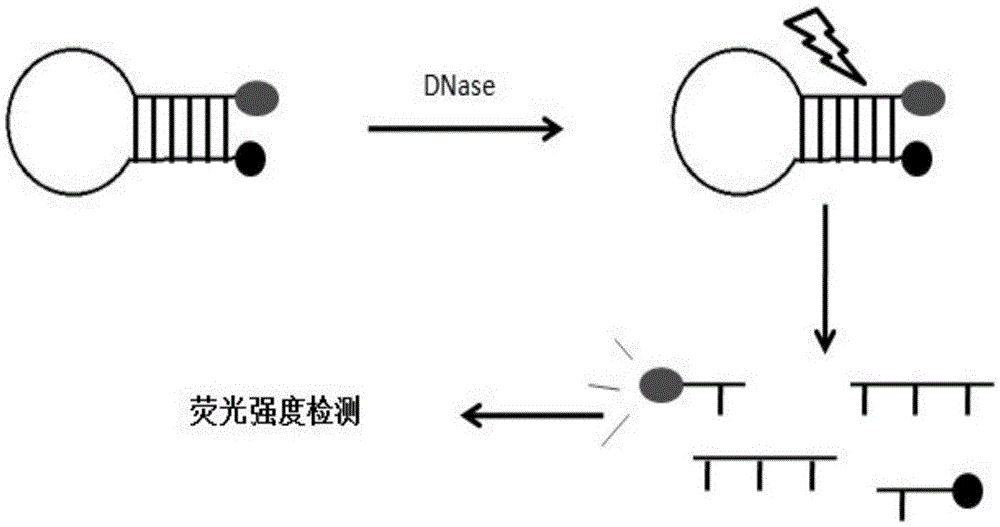
[0044] The design principles are as follows, see figure 1 .
[0045] 1) Palindromic complementary sequences with 5-15 bp at both ends (underlined part)
[0046] 2) In the middle is a 5-20nt DNA sequence (loop region)
[0047] 3) The probe can form a hairpin structure after annealing, and the Tm value of the two palindromic complementary parts is greater than 35°C; its stem structure includes at least one T, and its loop structure includes at least one A, one C, one T and 1G
[0048] 4) There are fluorescent groups and their corresponding quenching groups at both ends. The fluorescent group and its quenching group can be modified at both ends of the sequence or any DNA base in the sequence.
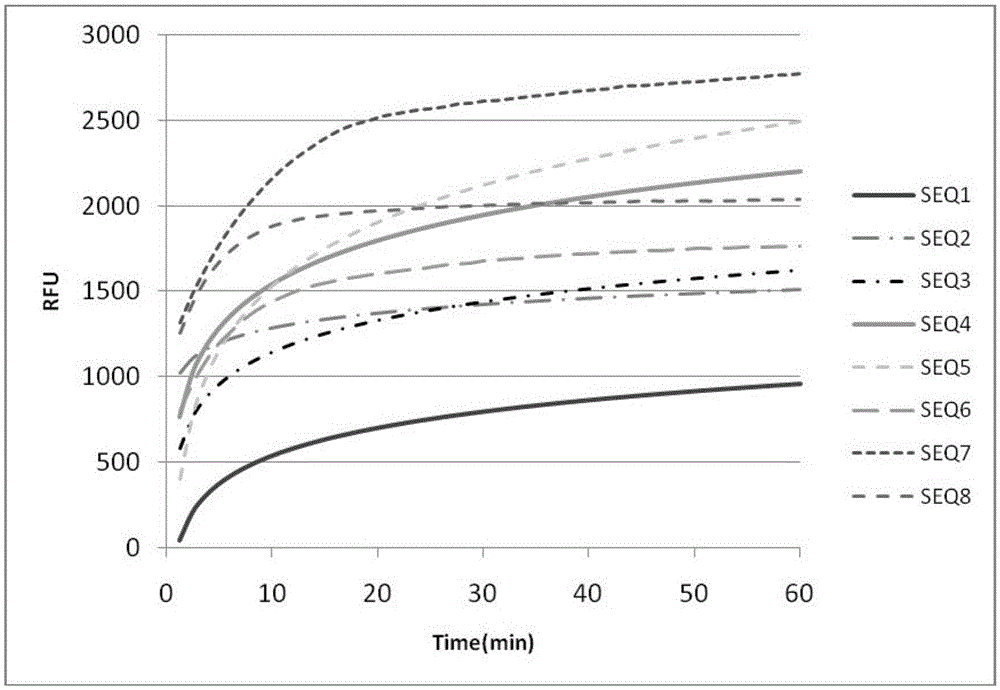
[0049] The sequences of SEQID.NO.1-8 (SEQ1-SEQ8 in the drawings) are as follows.
[0050]
[0051]
[0052] 2 Probe Synthesis
[0053] Entrust Shanghai Jierui Biotechnology Co., Ltd. to synthesize the ...
Embodiment 2
[0055] 1. Reagent synthesis and preparation
[0056] 1.1. DNasetestProbe
[0057] The probes are the 8 probes synthesized in Example 1.
[0058] 1.2. Preparation of DNaseTestProbe solution
[0059] Stock solution preparation (100pmol / ul): add sterilized Mini-Q water to each tube of DNaseTestprobe dry powder to a final concentration of 100pmol / ul, store at -20°C in the dark
[0060] Preparation of working solution (10pmol / ul): take 1.5ml tube without DNase, add 90ulddH 2 O and 10ul DNaseProbe stock solution, mix well, and store at -20°C in the dark.
[0061] 1.3. Reaction buffer
[0062] Formula: 500mMNaCl, 200mMKCl, 500mM Tris-HCl, 100mMMgCl 2 ,pH8.0
[0063] Prepare 10× reaction buffer solution with ultrapure water according to the above formula, sterilize under high temperature and high pressure, and store at -20°C.
[0064] 1.4. DNaseI positive samples
[0065] DNaseI (TAKARA), concentration 5U / ul. Dilute to 0.01U / ul with 1× reaction buffer.
[0066] 2. Methods an...
Embodiment 3
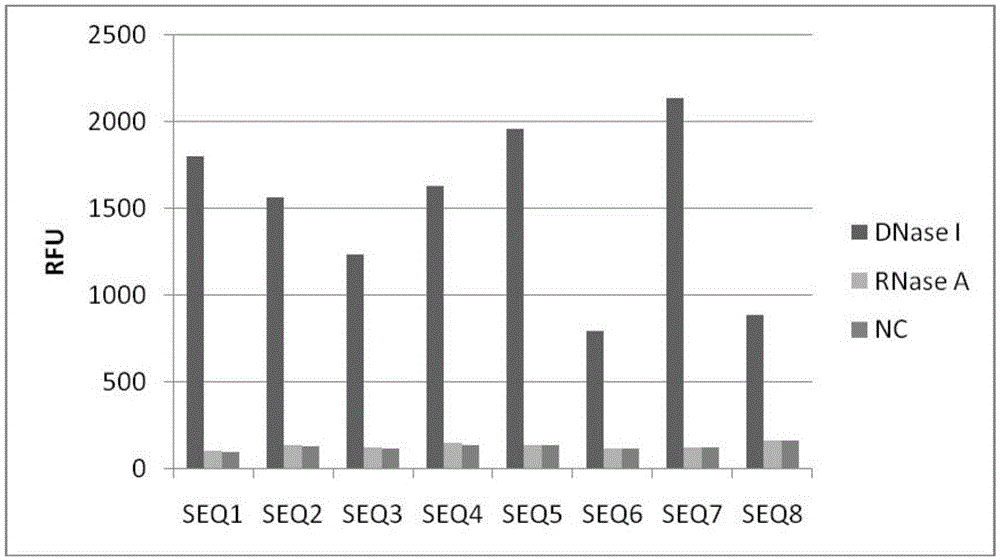
[0075] Example 3 Probe Specific Detection
[0076] 1. Reagent synthesis and preparation
[0077] 1.1. DNasetestProbe
[0078] The probes are the 8 probes synthesized in Example 1.
[0079] 1.2. Preparation of DNaseTestProbe solution
[0080] Preparation method is the same as embodiment two
[0081] 1.3. Reaction buffer
[0082] Preparation method is the same as embodiment two
[0083] 1.4. Samples
[0084] DNaseI (TAKARA), concentration 5U / ul. Dilute to 0.01 U / ul RNaseA (TianGen; #RT405-02; 10 mg / ml) with 1× reaction buffer and dilute to 1 ng / ul with 1× reaction buffer.
[0085] 2. Methods and steps
[0086] 2.1. Preparation of reaction system
[0087]
[0088]Mix the above components except the sample evenly, heat at 95°C for 3min, then 65°C to 25°C, drop 5°C every 30 seconds to anneal the probe. Take a white 8-section PCR tube, place it on a 96-well plate, add 9ul of reaction solution to each well, and then add 1ul of DNaseI (0.01U / ul). DNase-free ddH 2 O repl...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com