Liquid chip detection method for four types of pathogens of porcine virus diarrhea
A liquid-phase chip detection technology for porcine viral diarrhea, which is applied in the field of virology, can solve the problems of cumbersome operation, long experiment cycle, and no simultaneous detection, and achieve the effect of good repeatability and specificity
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
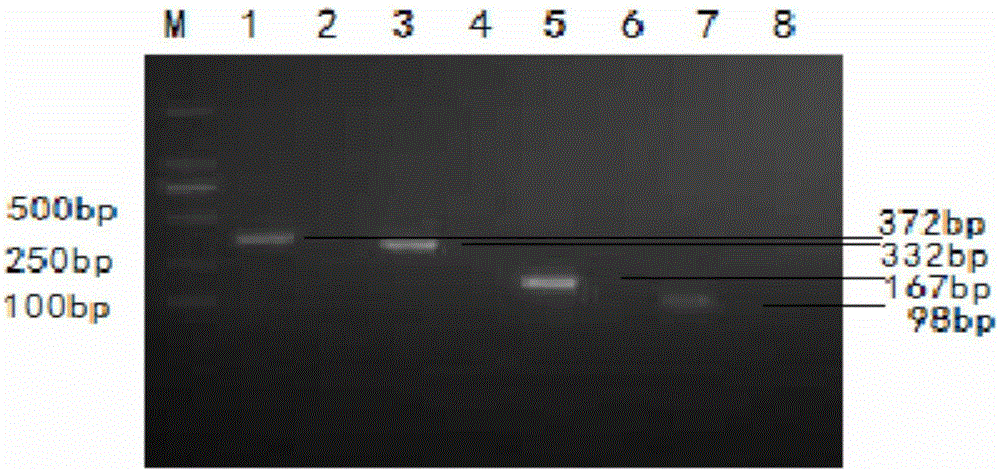
[0062] Design and synthesis of embodiment 1 primers and probes
[0063] Select the M gene of conservative PEDV on GENBANK (GenBank accession number is AF353511.1), the N gene of TGEV (GenBanK accession number is EU074218), the VP6 gene of PRoV (GenBank accession number is FJ617209.1) and pig Source BVDV's N pro After the gene sequence (GenBanK accession number is HQ258810.1) was analyzed online, DNASTAR software was used to analyze and design specific primers, probes and complementary probe chains for these genes. Amino (NH 2 (CH 2 ) 12 ) modification, all complementary probe strands and the 5' end of the downstream primers are labeled with biotin (Biotin), and the test screening is carried out after synthesis. The sequences of each viral primer and probe after screening are shown in Table 2. The probe and primer design of the present invention mainly pay attention to the following points:
[0064] Primer design principles: 1) The length of the general primer is 18-25 bas...
Embodiment 2
[0069] The preparation of embodiment 2 positive plasmid
[0070] Viral RNA extraction: Trizol lysis method was used to extract the RNA of the sample to be tested, and the operation steps were as follows: (1) Take 250 μl of each cytotoxicity and add 3 times the volume of Trizol lysate, mix with a vortex shaker, and stand on ice for 10 minutes; ( 2) Add 200 μl of chloroform, mix well with a vortex shaker, and let stand on ice for 10 minutes; (3) Centrifuge at 12000×g at 4°C for 15 minutes; (4) Take the supernatant, add an equal volume of isopropanol, and mix well. Stand on ice for at least 10 minutes, repeat step (3); (5) discard the supernatant, add 1ml of 75% absolute ethanol, mix upside down, repeat step (3), discard the liquid, blow dry, add 20 μl DEPC water, - Store at 80°C for later use.
[0071] Prepare cDNA by reverse transcription: 10ulRNA, 2ulprimemix, 0.5ulM-MLVRT, 0.5ulRRI, 5xReverseTranscriptaseM-MLVBuffer4ul, 2.5mMdNTP2ul, ddH2O1ul. 1h at 42°C, 15min at 75°C, 4, ...
Embodiment 3
[0081] Example 3 Microsphere coupled probe
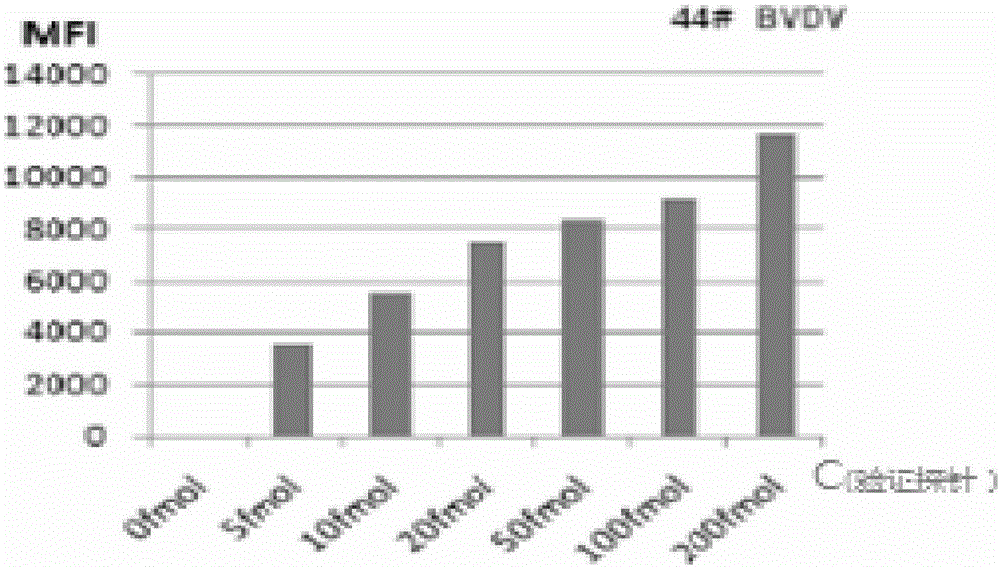
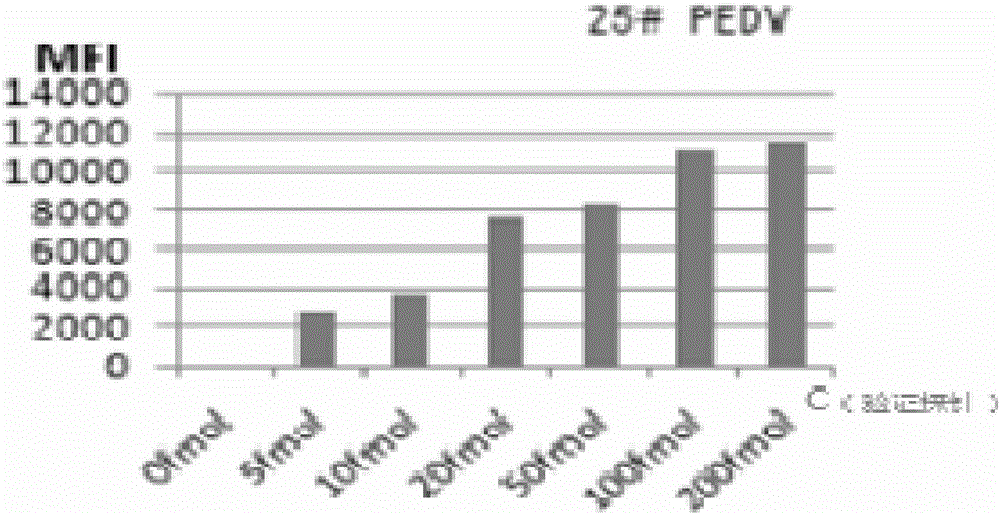
[0082]The probe BN-Probe of 44# microspheres coupled with BVDV; the probe PM1-Probe of 25# microspheres coupled with PEDV; the probe PVP6-Probe of 20# microspheres coupled with PRoV; the probe of 15# microspheres coupled with TGEV Probe TGEV-Probe, the coupling steps are as follows:
[0083] (1) Take 44# microspheres, 25# microspheres, 20# microspheres, and 15# microspheres (Luminex company) out of the 4-degree refrigerator and return to room temperature, vortex for 20 s, and sonicate for 20 s; take out 100 μl of microspheres Into a 1.5ml imported centrifuge tube;
[0084] (2) Centrifuge at 12000xg for 2 minutes, take out the centrifuge tube and put it on a magnetic stand, carefully remove the supernatant, resuspend the microspheres with 50 μl MES, vortex and shake to mix the microspheres completely;
[0085] (3) Add 10 μm probe BN-Probe 2 μl to 44# microspheres;
[0086] Add 10 μm probe PM1-Probe 2 μl to 25# microsphere;
[008...
PUM
| Property | Measurement | Unit |
|---|---|---|
| diameter | aaaaa | aaaaa |
Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com