Small interfering ribonucleic acid (siRNA) capable of interfering alpaca melanin cell CDK5 gene expression, expression plasmid and application thereof
A technology for melanocytes and expression plasmids, which is applied in the field of siRNA fragments and shRNA expression plasmids, and can solve the problems of lack of optimal and interfering siRNA
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0032] Design artificial siRNA sequences that interfere with alpaca CDK5 gene expression, and screen CDK5 siRNAs by cell transfection.
[0033] According to the full-length CDK5 sequence (HM623674) amplified from alpaca skin, and referring to the siRNA design principles, three siRNA double-stranded artificial sequences shown in Table 1 below were designed and screened.
[0034]
[0035] Inoculate 2 × 10 per well in a six-well tissue culture plate 5 Melanocytes, 2ml normal culture without antibiotics and fetal bovine serum, cultured in a carbon dioxide incubator for 18-24 hours, until the cells adhered to 60-80%. Prepare the liquid as follows: Solution A: Dilute 2-8µl siRNA duplex with 100µl siRNA transfection medium sc-36868 for each transfection; Solution B: Dilute 2-8µl siRNA transfection reagent sc-29528 with 100µl for each transfection siRNA transfection medium sc-36868 diluted.
[0036] Add the siRNA duplex (solution A) directly into the diluted transfection reagent ...
Embodiment 2
[0043] According to the CDK5 siRNA sequence screened in Example 1, the shRNA with the sense strand and antisense strand sequence shown in Seq ID No.3 and Seq ID No.4 is designed, and the shRNA is identified as Seq ID No.1 and Seq ID No. The 2 sequence is used as the stem structure of shRNA, and CGAA and UUCG are used as the hairpin structure between the two, thus forming the shRNA structure:
[0044] Sense Strand: 5’-CACC GCGACAAGAAGCUGACUUU CGAA AAAGUCAGCUUCUUGUCGC -3'
[0045] Antisense strand: 5'-AAAA GCGACAAGAAGCUGACUUUUUCG AAAGUCAGCUUCUUGUCGC -3'.
[0046] According to the above shRNA sequence, a pair of shRNA oligomeric single-stranded DNA was synthesized, and a negative control sequence was attached. The specific sequence is shown in Table 2.
[0047]
[0048] Annealing of double-stranded DNA: use ddH to synthesize 1 pair of oligomeric single-stranded DNA 2 O was dissolved to 100 µM, 5 µl of each complementary single strand was mixed, and annealed according t...
Embodiment 3
[0064] The alpaca CDK5 pENTR / U6 shRNA expression plasmid was used to transfect melanocytes through liposomes, and the biological effects of melanocytes were analyzed.
[0065] During transfection, take recovered 1×10 5 Melanocytes were seeded in six-well plates and cultured in melanocyte medium (Sciencell, San Diego, USA) for 24 hours, and adhered to 70%. On the second day, the medium was discarded, and 800 µL of serum-free medium was added, and the complex of DNA fectin Transfection Reagent (TIANGEN BIOTECH, BEIJING CO. LTD) and alpaca CDK5 pENTR / U6 shRNA was added and mixed, and incubated at 37°C for 20 Hour. Remove the transfection reagent, add 20ml complete medium containing serum, and culture at 37°C for 3 days. Cells were collected for RNA extraction. Each experimental group has 3 repetitions.
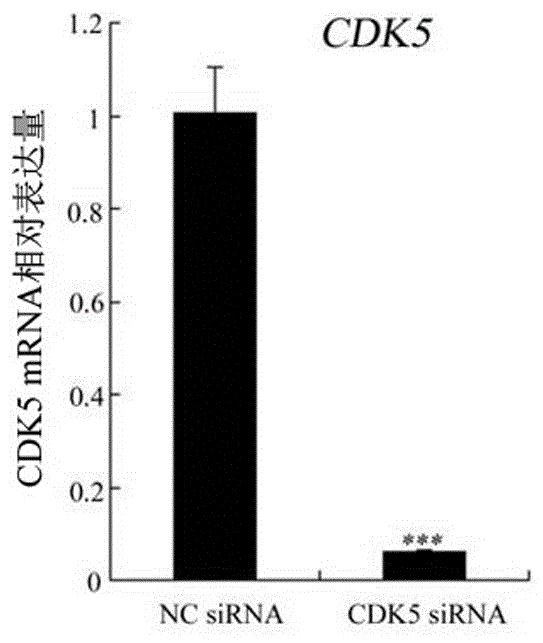
[0066] Real-time fluorescent quantitative PCR experiments were performed to detect changes in the expression of target genes.
[0067] The primers required are as follows: ...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com