ApoE kit, primers and use thereof
A kit, the technology of use, applied in the field of biology
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
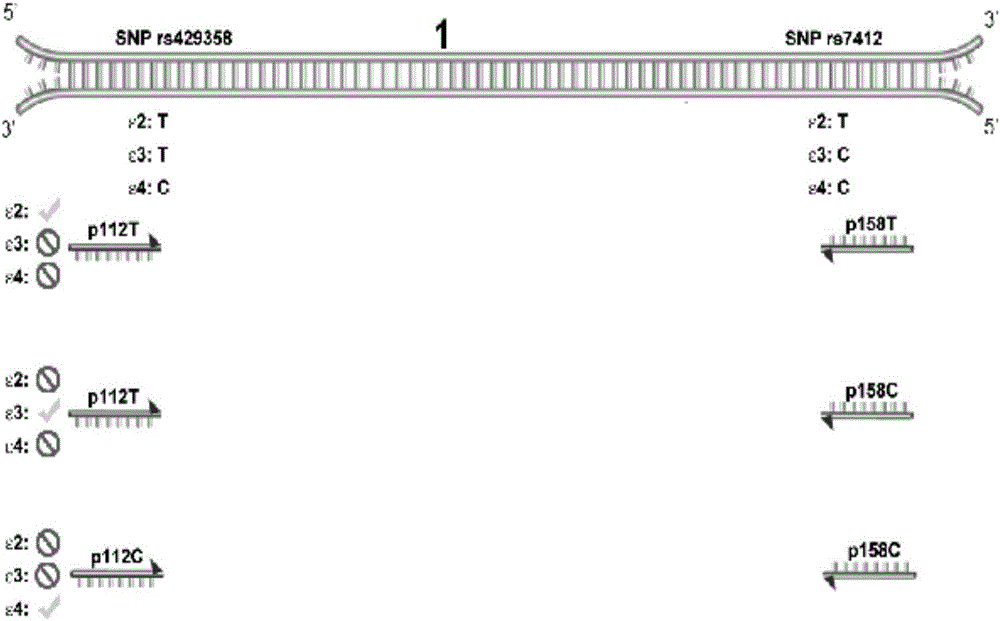
[0324] In the example, as figure 1As shown, the primer p112T only matches the nucleic acid sequence of SNP rs429358 being T, the primer p112C only matches the nucleic acid sequence of SNP rs429358 being C, the primer p158T only matches the nucleic acid sequence of SNP rs7412 being T, and the primer p158C only matches the nucleic acid sequence of SNP rs7412 being C nucleic acid sequence match. Therefore, the PCR system containing the primer pair p112T / p158T can only amplify the ApoE allele (ie ε2) in which SNP rs429358 is T and SNP rs7412 is T; the PCR system containing the primer pair p112T / p158C can only amplify SNP rs429358 The ApoE allele with T and SNP rs7412 as C (namely ε3); the PCR system containing the primer pair p112C / p158C can only amplify the ApoE allele with SNP rs429358 as C and SNP rs7412 as C (ie ε4). Therefore, using the above three primer pairs, different alleles of ApoE can be specifically amplified, and the type of human ApoE alleles in the DNA sample can ...
Embodiment 2
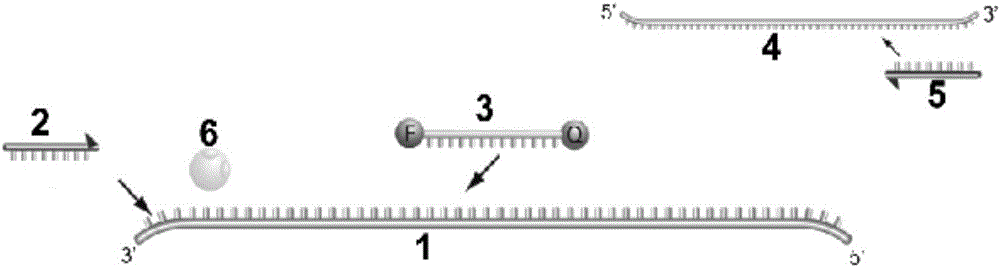
[0326] In the example, as figure 2 As shown, the two sections of the DNA molecular probe 3 detached from the fluorophore (F) are coupled with a fluorophore and a quenching group respectively. At this time, since the quenching group can absorb the excitation energy of the fluorophore, therefore, Only background fluorescence signal can be detected in the system. The DNA molecular probe 3 detached from the fluorophore (F) can specifically complement and match the antisense strand 1 of the DNA template of the ApoE gene, and the matching sequence is located between the DNA sequences corresponding to the forward amplification primer 2 and the reverse amplification primer 5 , after the PCR expansion reaction starts, forward amplification primer 2 and the DNA molecule probe 3 that fluorophore (F) breaks off will combine with ApoE gene DNA template antisense strand 1, and reverse amplification primer 5 will combine with ApoE gene The sense strand 4 of the DNA template binds, and unde...
Embodiment 3
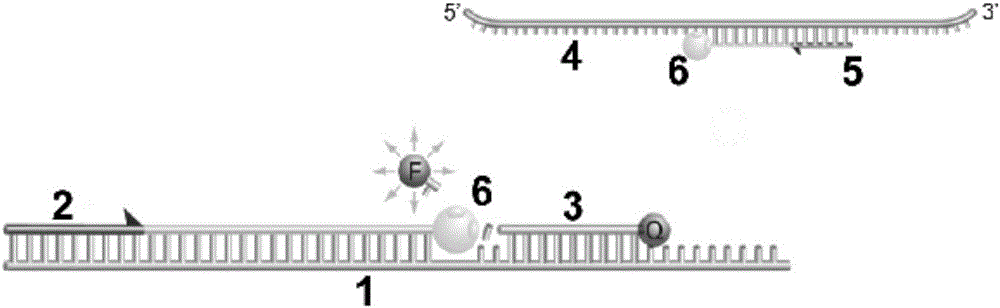
[0328] In the example, as image 3 As shown, DNA polymerase 6 starts from the forward amplification primer 2 or the reverse amplification primer 5 during the PCR reaction, using the antisense strand 1 of the ApoE gene DNA template or the sense strand 4 of the ApoE gene DNA template as a template, and starts The extension of DNA, when DNA polymerase 6 encounters the DNA molecular probe 3 that is bound to the fluorescent group (F) on the antisense strand 1 of the ApoE gene DNA template in the process of mediating DNA extension, DNA polymerase 6 will The 5' end of the DNA molecular probe 3, which is detached from the fluorescent group (F) by its exonuclease activity, cuts off the nucleotides on the probe one by one, causing the fluorophore to detach from the probe and combine with the quenching group After separation, the fluorescent signal excited by the fluorophore can be detected at this time, and thus it can be determined that the detection sample contains the ApoE allele typ...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com