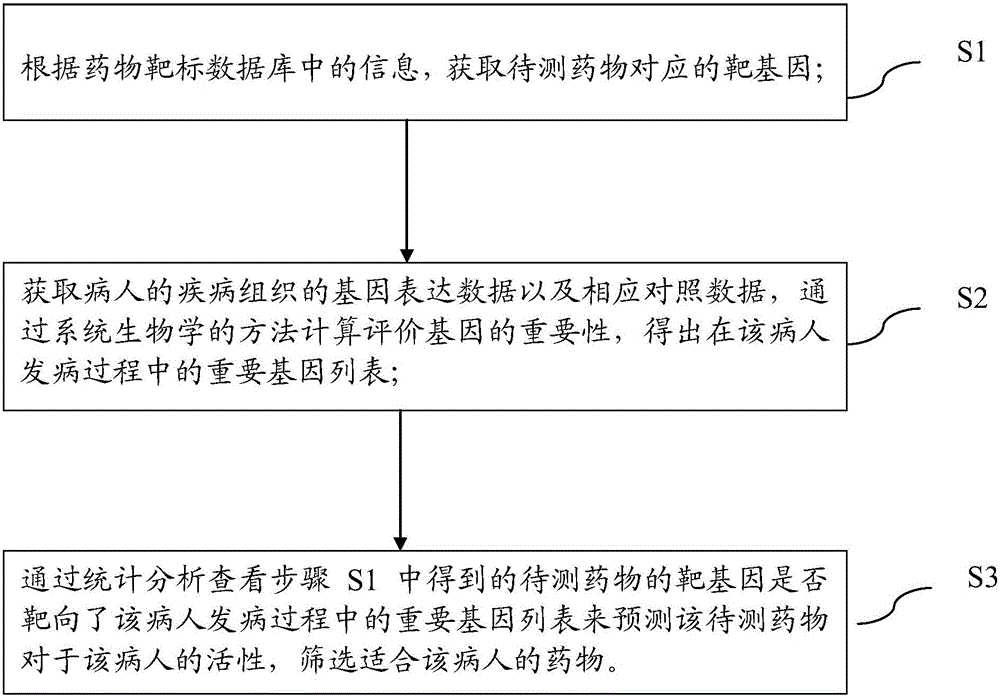
Pharmaceutical activity prediction and selection method based on genetic expressions and drug targets
A technology of drug activity and screening method, applied in the field of biomedicine, can solve the problems of overfitting, not considering biological mechanism, affecting practical value, etc., and achieve the effect of easy use, wide application range and high efficiency
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
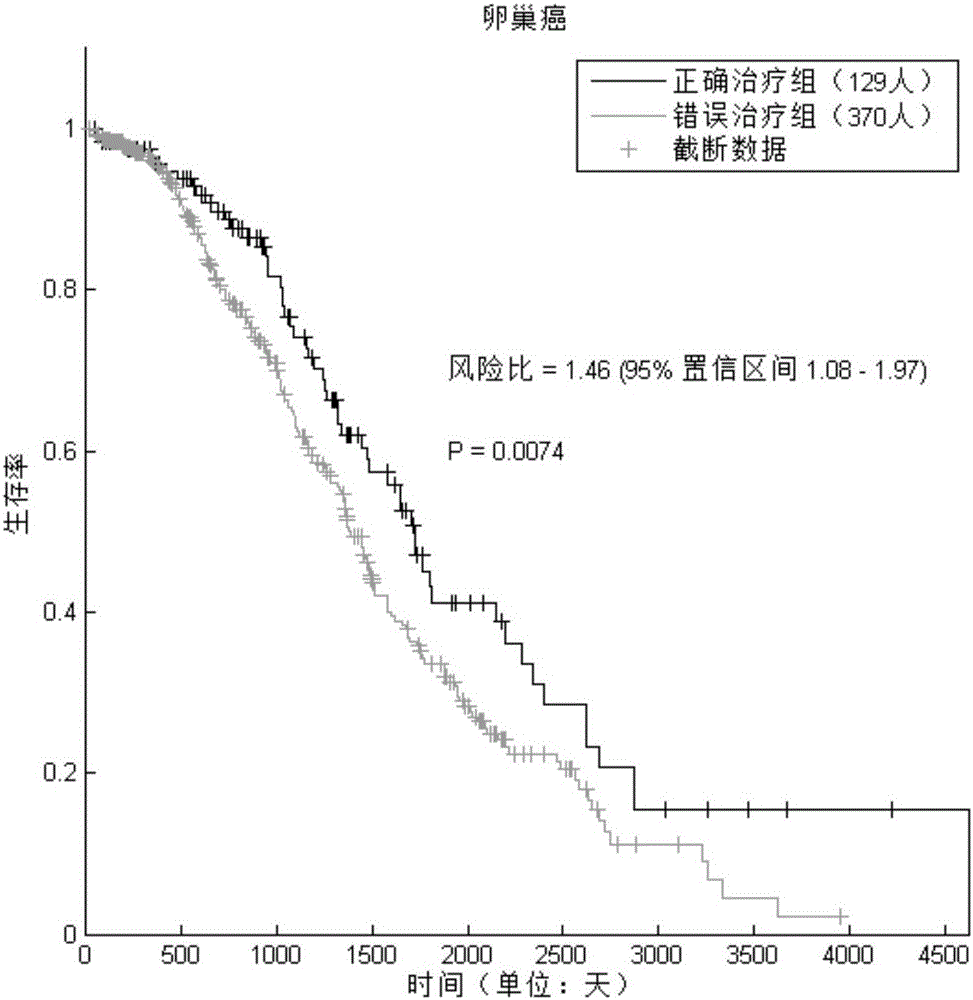
[0043] Using the method of the present invention to predict the drug activity of ovarian cancer patients
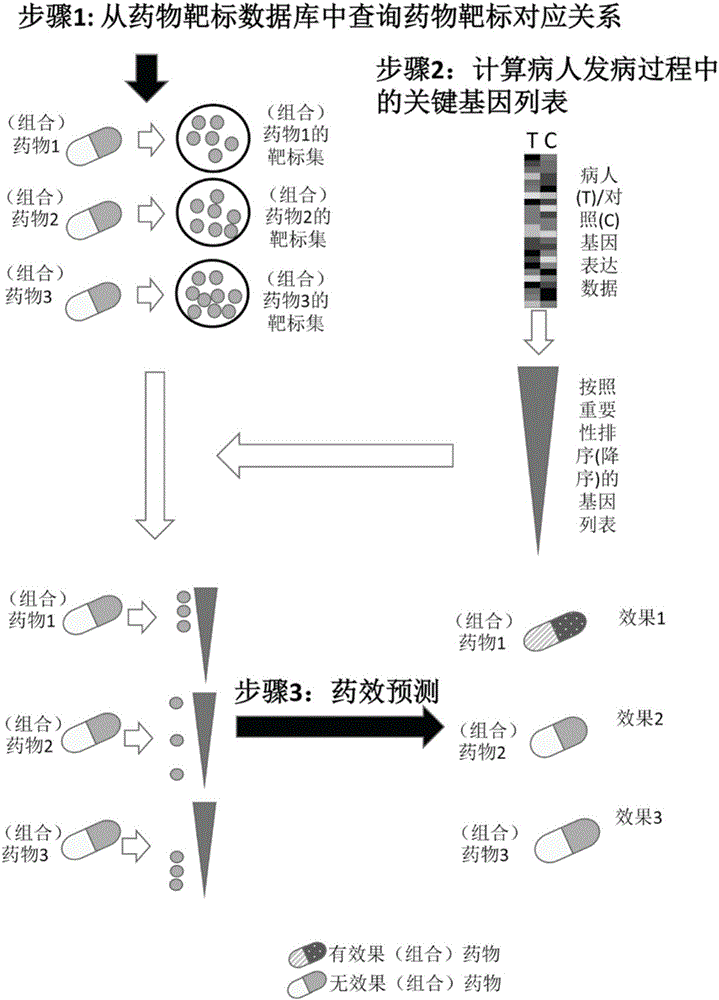
[0044] 1. Collect human successfully marketed or researched drugs and their targets
[0045] Find drug target databases (including DGIdb: http: / / dgidb.genome.wustl.edu / , DrugBank: http: / / www.drugbank.ca / and TTD: http: / / bidd.nus.edu.sg / group / ttd / ttd.asp), to get all the drugs contained in the database and their corresponding target data.
[0046] 2. Collection of gene expression data and clinical data of ovarian cancer patient samples
[0047] The gene expression data (level 3 data of AgilentG4502A chip), the patient's prognosis tracking data (death time, death status) and the patient's medication information of ovarian cancer (OV) patients and control samples were downloaded from TCGA (The Cancer Genome Atlas). Excluding samples with lack of drug information (or unknown drug targets) and missing prognostic information, a total of 499 cancer patients and 10 normal ov...
Embodiment 2
[0056] Prediction of drug activity in patients with glioblastoma multiforme using the method of the invention
[0057] The steps of this embodiment are the same as embodiment 1, and the other steps are as follows:
[0058] 2. Collection of gene expression data and clinical data of glioblastoma multiforme patient samples
[0059] Download the gene expression data (level 3 data of AgilentG4502A chip) of glioblastoma multiforme patients (GBM) and control samples from TCGA (The Cancer Genome Atlas), the patient's prognosis tracking data (death time, death status) and Patient medication information. Excluding samples with lack of drug information (or unknown drug targets) and missing prognostic information, a total of 193 cancer patients and 10 corresponding normal tissue samples were obtained. Gene expression data, prognosis tracking information and drug information of these 193 patients (445 drug groups information).
[0060] 3. Calculating the key gene list in the patient's...
Embodiment 3
[0068] Using the method of the present invention to predict the activity of combined drugs for breast cancer patients
[0069] 1. The drug target database used in this embodiment is the same as in Example 1 and Example 2. For the combined drug, the target is the union of the target data of these drugs, and the other steps are as follows:
[0070] 2. Download the two breast cancer datasets MDA1 and MDA / MAQC-II from NCBI GEO. The data set contains the gene expression data of these patients (GPL96 microarray data of 278 breast cancer patients) and the response status information after subsequent T-FAC (paclitaxel, fluorouracil, doxorubicin and cylclophosphamide) treatment. Download the GSE9574 dataset from NCBI GEO, which contains gene chip data (GPL96 chip) of normal breast tissue. Firstly, the gene chip probe data of breast cancer patients and control data are matched to the gene ID, and the gene expression data with the gene ID as the basic unit is obtained through processi...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com