Rice disease resistance-related gene osdr11 and its application in rice disease resistance
A disease-resistance-related gene and rice technology, applied in the field of genetic engineering, can solve the problems of low plant disease resistance and environmental pollution
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
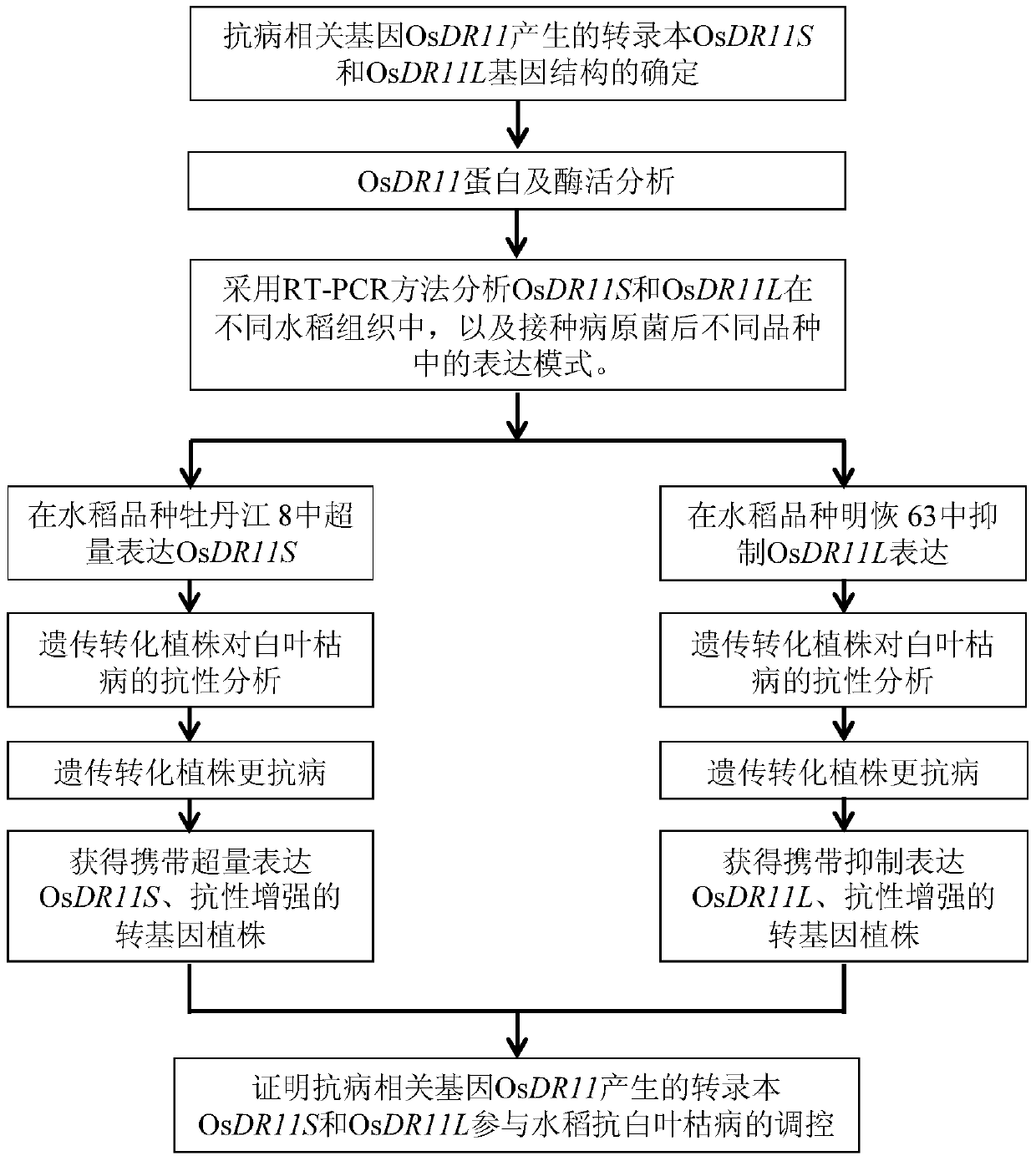
[0026] Example 1: Isolation and clone OsDR11 gene and gene structure analysis
[0027] 1. Determination of two different transcripts of OsDR11
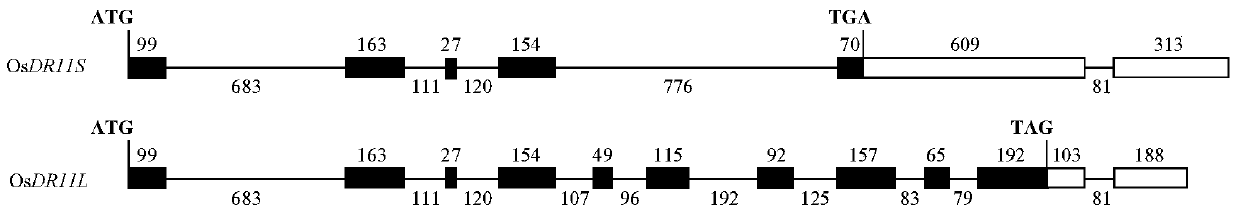
[0028] In the previous research work of the present invention, the State Key Laboratory of Crop Genetic Improvement, where the inventor works, used cDNA chip technology to analyze and found that the cDNA clone EI39C8 derived from rice variety Minghui 63 was inoculated with Xanthobacterium bacillus in rice variety Minghui 63. The expression level was induced to increase 2.4-2.6 times (Zhou et al., 2002). The inventors of the present invention sequenced the insert fragment of the EI39C8 clone, and used the EI39C8 sequence as a template to analyze the insert sequence of EI39C08 by BLASTN method, and found that the encoded product was homologous to Arabidopsis LAMMER protein kinase. According to the prediction results of software such as GenScan, FGENESH (http: / / www.softberry.com) and ORF Finder (http: / / www.ncbi.nlm.nih.gov / gorf / gorf.htm...
Embodiment 2
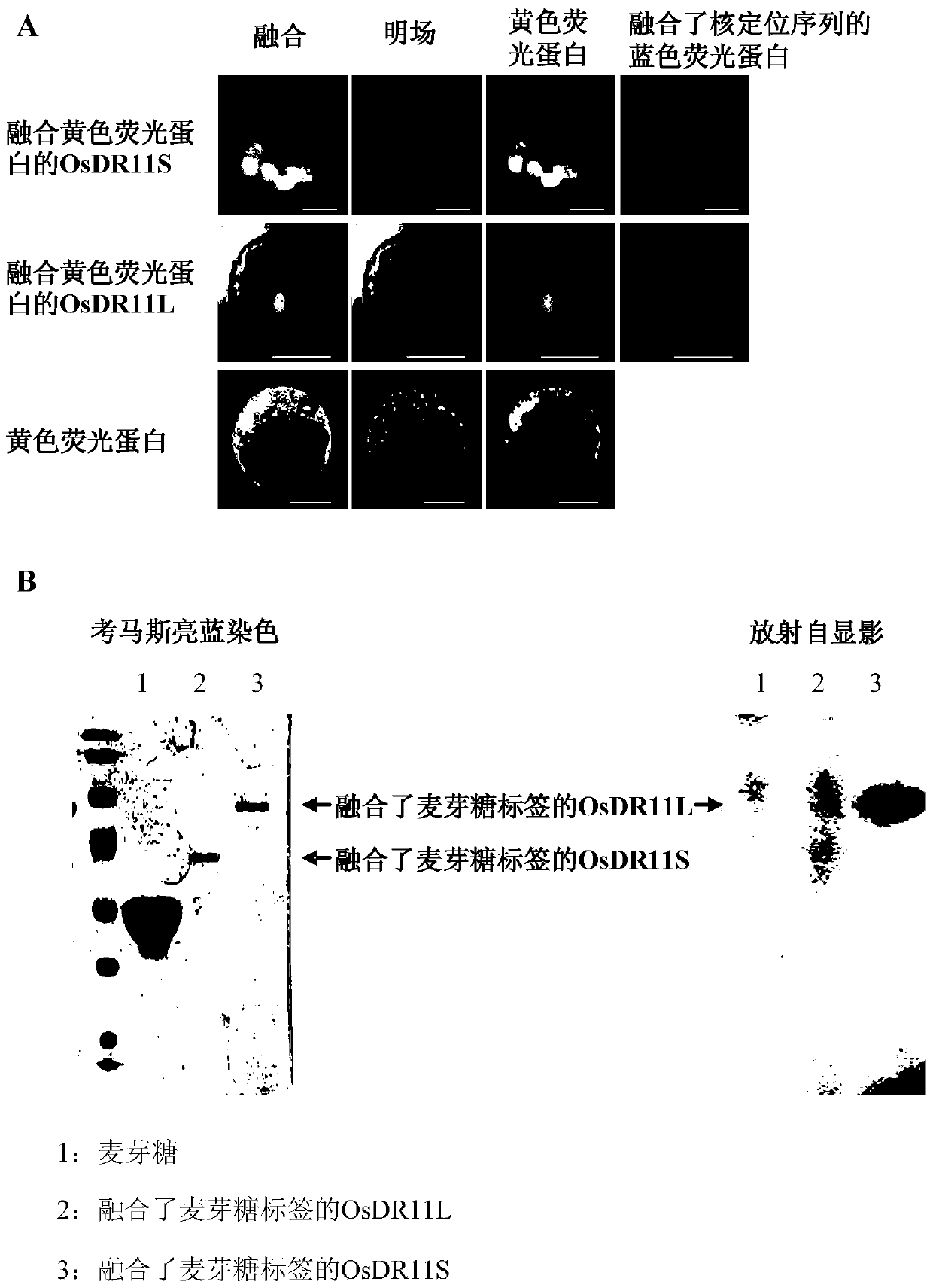
[0034] Embodiment 2: Analysis of OsDR11 gene coding product
[0035] 1. Analysis of subcellular localization of OsDR11 gene-encoded product
[0036]The amino-terminal of OsDR11 has the same nuclear localization sequence as other family members, indicating that both OsDR11S and OsDR11L may be localized in the nucleus to play a role. Clone EI39C8 with cDNA, use 39C8-4F4 (5′-ATTCTAGAATGGAGTGCTTGGCCGAGAT-3′) and 39R-XBA1 (5′-CTTCTAGACCTTAATAGCACTGGACTTG-3′) to amplify the full-length cDNA of OsDR11S, and connect it to the pM999-YFP vector after single digestion with XbaI Above, pick positive monoclonal sequencing verification, construct OsDR11S subcellular localization vector. Using the cDNA clone EI114F3 as a template, use 39C8-4F4 (5′-ATTCTAGAATGGAGTGCTTGGCCGAGAT-3′) and F3R-XBA1 (5′-CTTCTAGACATACAAGCAACAAATGAGC-3′) to amplify the exogenous fragment by PCR, and then ligate it to pM999 after single digestion with XbaⅠ -On the YFP vector, pick the positive monoclonal sequencing ...
Embodiment 3
[0042] Example 3: Analysis of the expression pattern of the OsDR11 gene
[0043] 1. Analysis of the expression pattern of two different transcripts of OsDR11 in rice tissues
[0044] RNA samples from different tissues of indica rice variety Minghui 63 at different stages, including callus, roots and leaves at two tillering stages, leaves, leaf sheaths and young panicles at booting stage, flag leaves and panicles at heading stage, and at flowering stage Stamens and pistils were detected by real-time quantitative PCR after reverse transcription (see Figure 4 in A panel). The experimental results confirmed that the two transcripts were expressed in different tissues at different growth stages of rice, with higher expression in leaves, leaf sheaths at the booting stage and gametes at the flowering stage, and OsDR11S was expressed in mature vegetative organs such as leaves and leaf sheaths OsDR11L is more abundant than OsDR11S in young vegetative organs such as callus and reprod...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com