High efficiency clustering method based on locality-sensitive hashing and non-parametric Bayes method
A local sensitive hashing, non-parametric technology, applied in the direction of electrical digital data processing, special data processing applications, instruments, etc., can solve the problems of inaccurate greedy algorithm results, results dependent on sequence input order, etc., to achieve high accuracy , estimated accurate effect
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
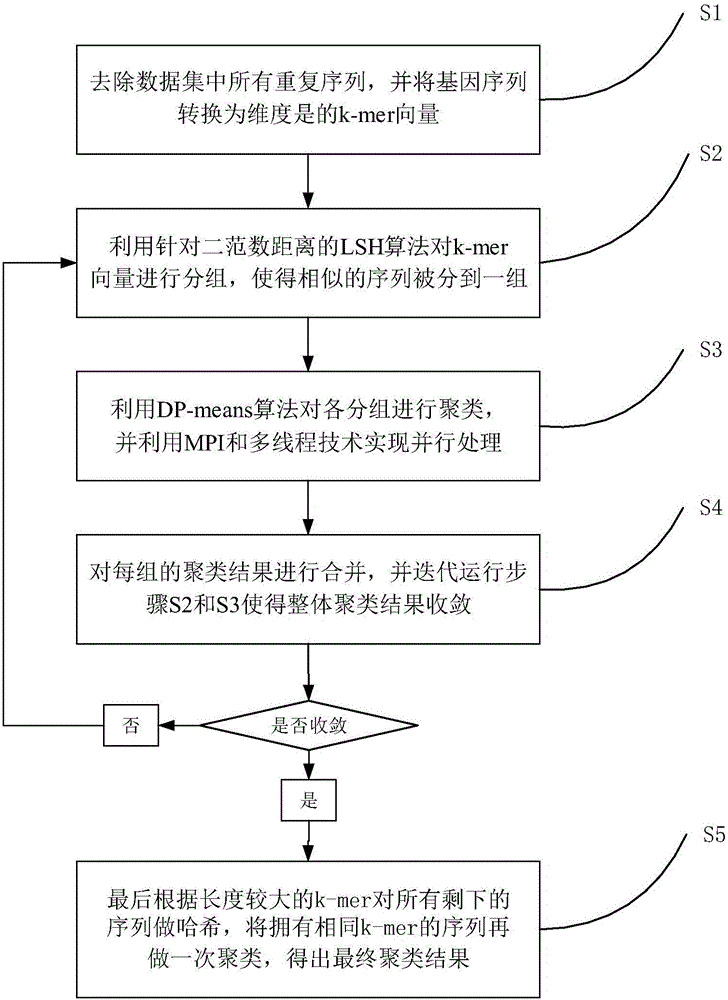
[0034] The clustering algorithm based on locality-sensitive hashing (LSH) and non-parametric Bayesian method (DP-means) involved in this embodiment will be described in detail below. Core algorithm pseudocode of the present invention sees figure 1 ; The flow chart of the clustering algorithm based on locality-sensitive hashing and non-parametric Bayesian method proposed by the present invention is shown in figure 2 .
[0035] S1. Remove all repetitive sequences in the data set, and convert the gene sequence to a dimension of 4 K The k-mer count vector
[0036] Since there may be a large number of identical gene sequences in general metagenomic 16s rRNA samples, the abundance of a microorganism can be known through the number of these sequences. At the same time, in a cluster, the sequence with higher abundance is more likely to be a cluster. class center. Therefore, we set a weight for each sequence, and the initialization of the weight is the number of repeated sequences...
Embodiment 2
[0056] Example 2 Cluster Analysis of Simulation 16s RNA Gene Data Set
[0057] In order to compare the clustering results of the algorithm with the ground truth of the data set, we conducted comparative experiments on the simulation data set. The simulation data set is generated by the software Grinder. There are 5 groups in total. The parameters of each group are shown in Table 1 below:
[0058] Table 1 Simulation data set generation parameters
[0059]
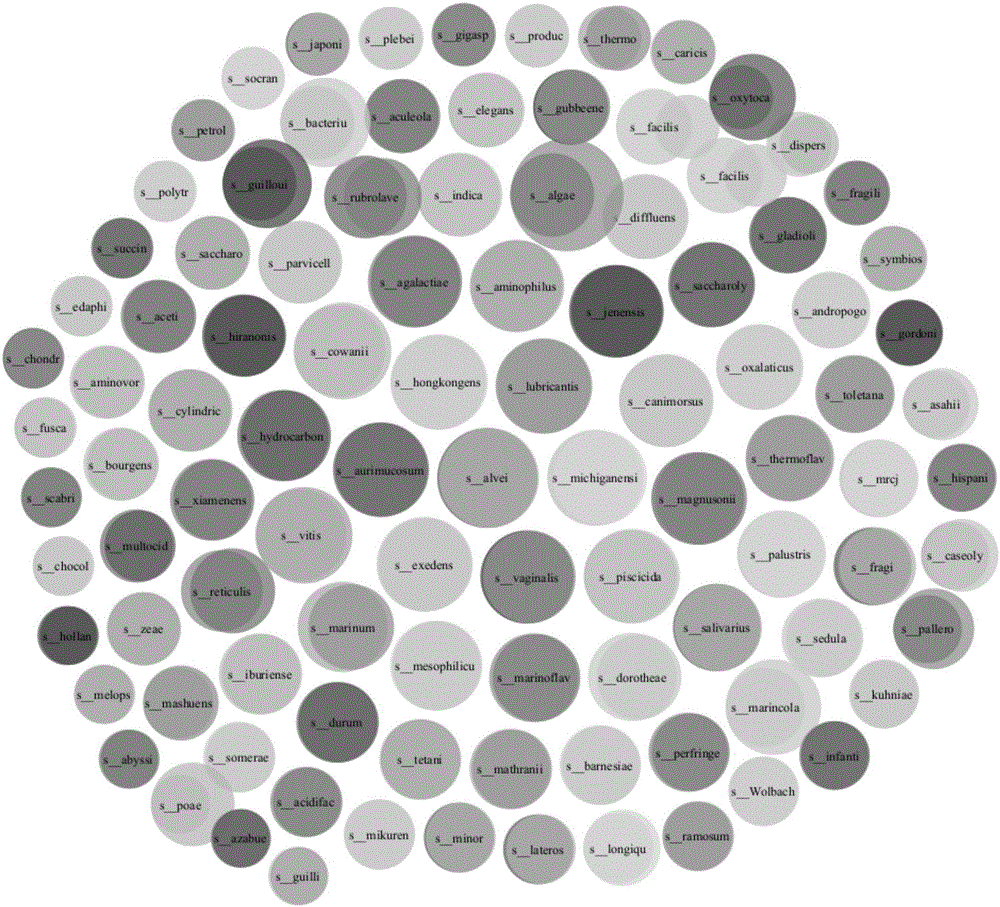
[0060] The visualization result of the clustering result on Sim5 by the method of embodiment 1 is as follows Figure 3-A shown. Figure 3-A The area of each circle in is proportional to the size of the cluster (the number of sequences in the cluster), the left of each pair of circles represents the ground truth, the right represents the clustering result of this algorithm, and the overlapping area represents both intersection of . This illustration shows the comparison results of the largest 60 clusters in Sim5 (acc...
Embodiment 3
[0061] Example 3 Cluster Analysis of Taihu Lake Microbial 16s rRNA Metagenome Data Set
[0062] This data set is a 16s rRNA metagenomic data set collected for the study of water pollution in Taihu Lake. It contains 81 water surface microbial samples collected in 9 different months in 2012. The entire dataset contains 316,153,464 original sequences, the sequence length is 80bp, and the file size is 30GB.
[0063] Figure 4-A It shows the time required for the method (DACE) of Embodiment 1 to process various data scales under different numbers of CPU cores. It can be seen that by using the message passing interface and multi-threading technology, the algorithm (DACE) of Embodiment 1 can effectively utilize computing resources, and greatly reduce the running time when the number of CPU cores increases. In particular, when the data size is large, the running time can be proportionally reduced as the number of CPU cores increases. It can be seen that the scalability (Scalability...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com