Capture library for detecting 50 tumor-relevant hotspot mutation genes and kit
A technology related to mutant genes and tumors, which is applied in the field of sequence capture library preparation, can solve the problems of non-specific amplification of some regions that cannot be amplified, the high cost of molecular probe synthesis, and poor uniformity, so as to increase the amount of effective data and high Cost-effective, ensuring consistent results
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
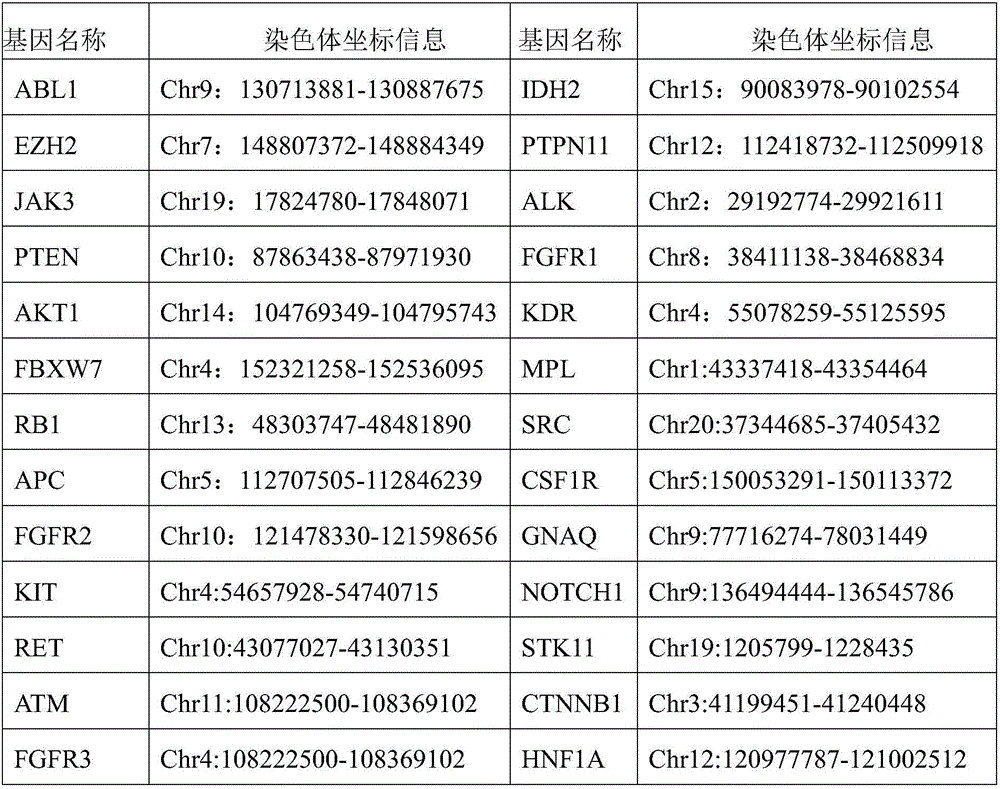
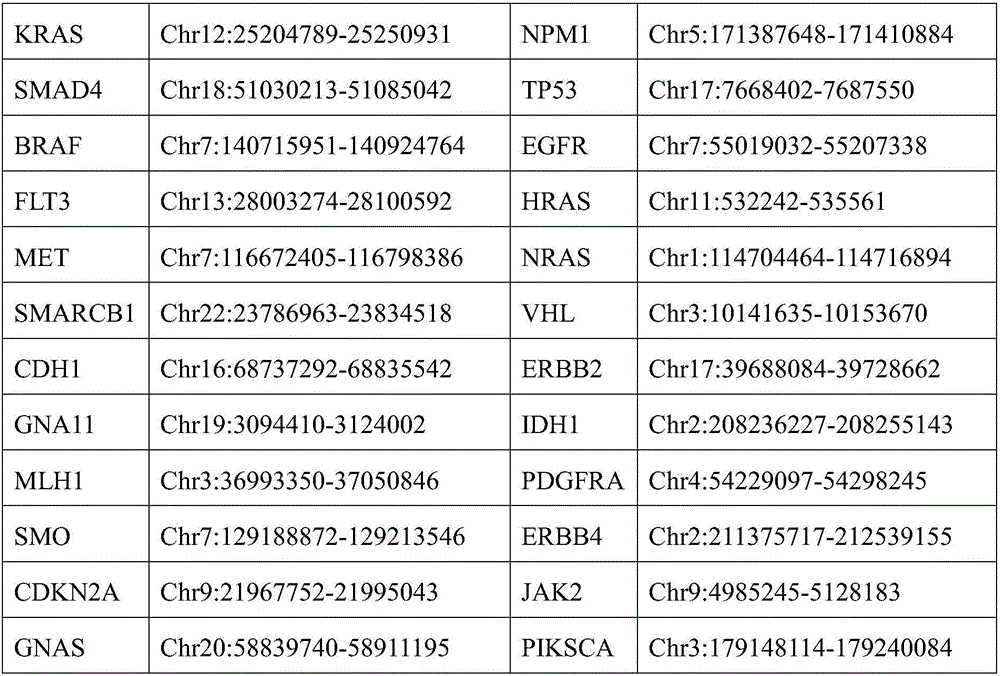
[0036] Specifically, on the basis of the present invention based on the existing or obtained information on 50 hot-spot mutation genes related to tumors, a detection kit for 207 hypermutated regions of 50 hot-spot mutation genes related to tumors was constructed. The kit first The extracted sample DNA is ultrasonically fragmented into 200-250bp DNA fragments (but not limited to the ultrasonic fragmentation method), and the DNA fragments are connected to the index adapter through end repair, adding "A" and ligation, and then use the constructed The capture library is hybridized with the DNA fragments containing the target gene region to be captured, and the DNA fragments of the tumor-associated hotspot mutation genes are captured. These DNA fragments are sequenced, and the sample detection results are obtained after bioinformatics data analysis. The technical solution includes the entire experimental process from sample genomic DNA to the output of sequencing results, mainly inc...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com