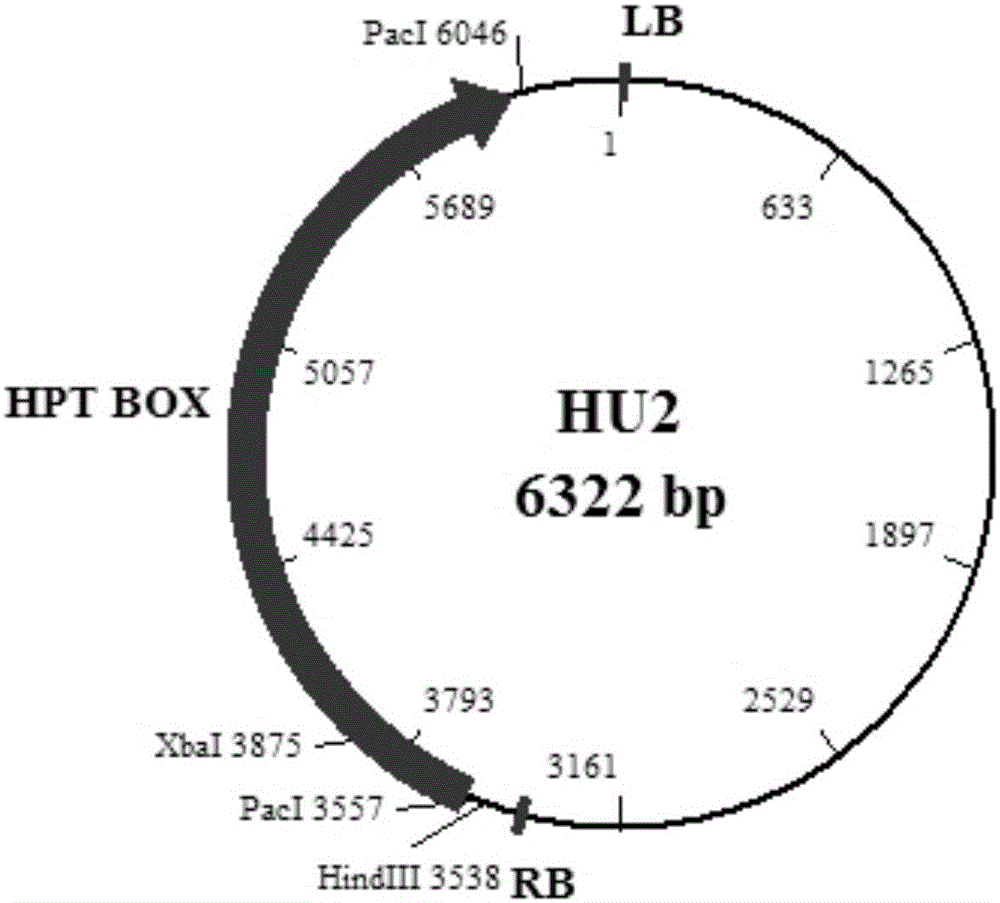
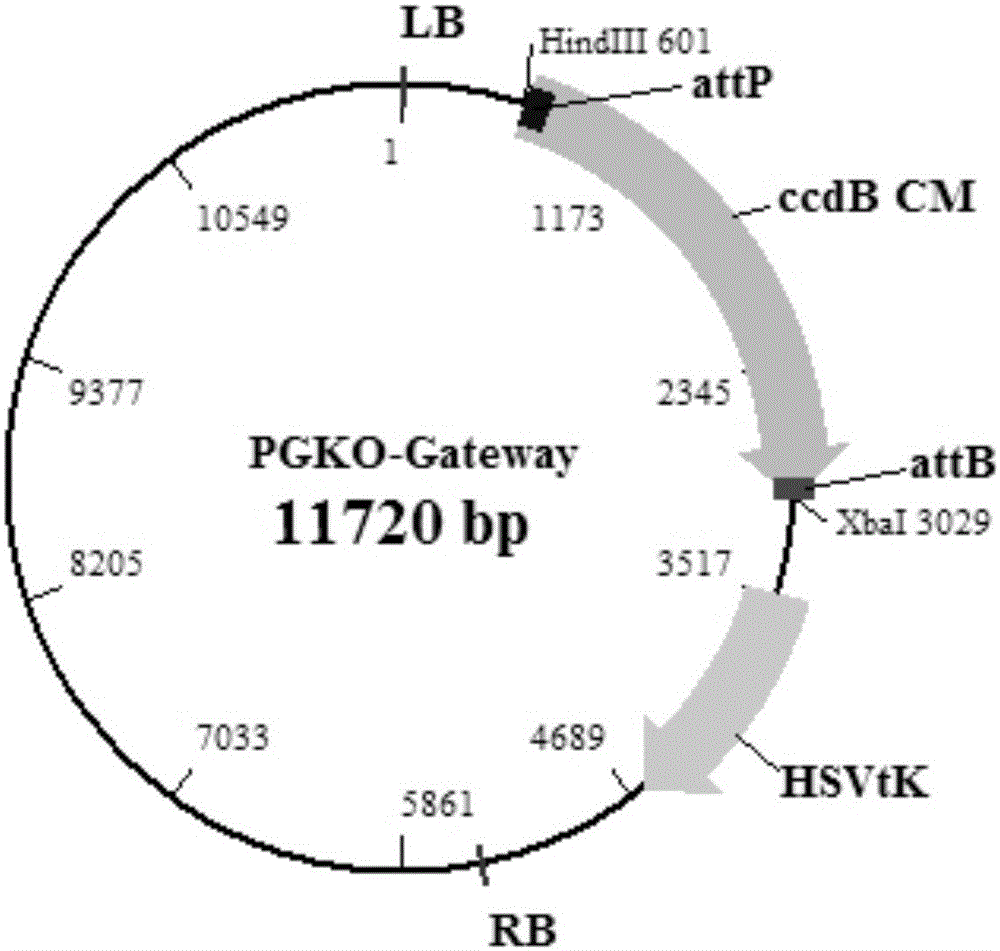
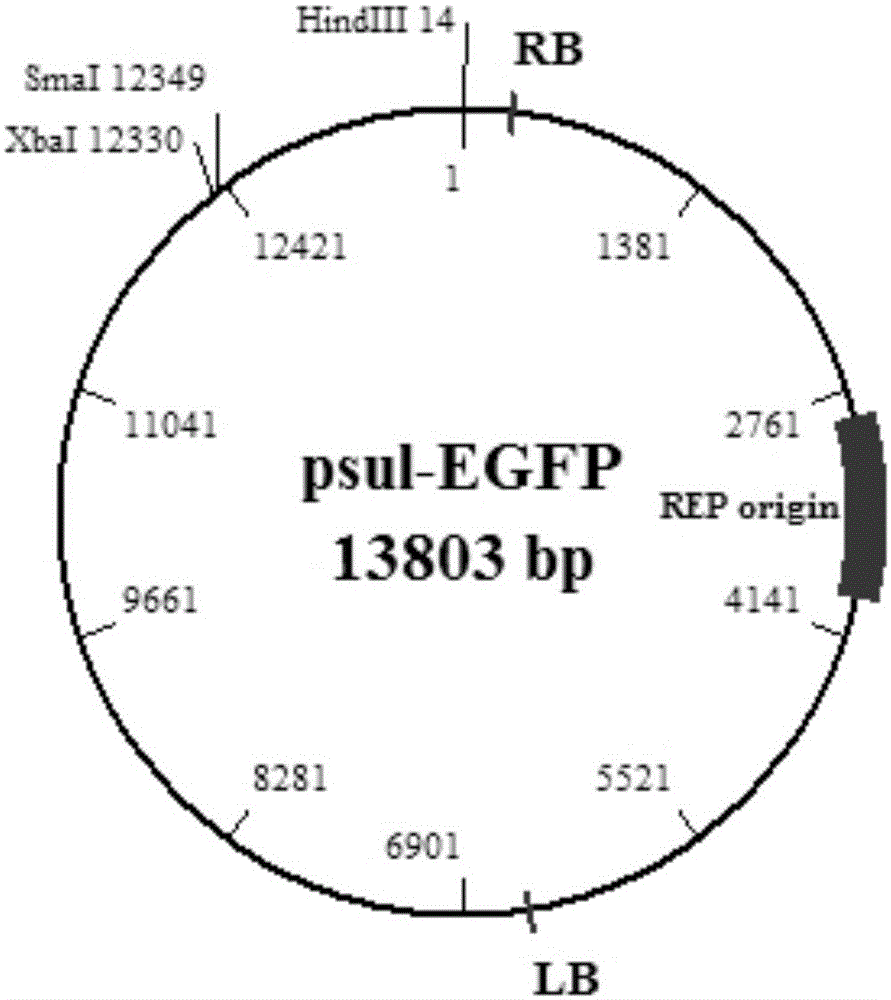
Efficient gene knockout vector construction method and constructed engineering bacteria
A gene knockout and construction method technology, applied in the biological field, can solve the problems of cumbersome positive transformants, etc., and achieve the effect of shortening the research cycle and speeding up the research
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0047] The construction of embodiment 1 recombinant vector
[0048] 1. The vector PGKO-Gateway (Methods in Molecular Biology, vol.344: Agrobacterium Protocols, 2 / e, volume 2) (20 years from the date of application can be obtained from the Institute of Microbiology, Chinese Academy of Sciences) was carried out through Xba Ⅰ and Hind Ⅲ Double enzyme digestion (see electrophoresis after enzyme digestion Figure 5 ), recover the large fragment of the vector after digestion. The reagents used for enzyme digestion are listed in Table 1.
[0049] Table 1
[0050] 10×RE Buffer 5μL Xba I 1μL HindⅢ 1μL PGKO-Gateway 10μL wxya 2 o
Make up 50μL
[0051] 2. The vector Psul-EGFP was digested by Xba Ⅰ and Hind Ⅲ (see the electrophoresis diagram after digestion). Image 6 ), recover the short fragments after enzyme digestion. The reagents used for enzyme digestion are listed in Table 2.
[0052] Table 2
[0053] 10×RE Buffer 5μ...
Embodiment 2
[0074] Example 2 Application and Verification of Recombinant Vectors in Gene Knockout
[0075] 2.1 Linearization of PGKO-HPT vector
[0076] Table 8
[0077] 10×cutSmart Buffer 30μL Pac I 10μL PKGO-HPT -μL (take 20-30μg)
[0078] wxya 2 o
Make up 300μL
[0079] The PGKO-HPT carrier prepared in Example 1 was digested overnight at 37°C. The reagents used for the digestion were shown in Table 8. The next morning, 4ul of PacI (New England Biolabs, Cat.no.R0547) and 4ul of Nt.BbvCI (New England Biolabs , Cat.no.R0632), and then placed in a 37°C water bath for 2 hours of enzyme digestion, and then 5ul was taken to run the gel to identify the enzyme digestion efficiency. All digested products were recovered with the SanPrep column DNA Gel Extraction Kit (Lot: 14023036Y) of Sangon Bioengineering (Shanghai) Co., Ltd., and the content was determined with a NanoDropND-1000 UV-Vis spectrophotometer after recovery. Determination. Aliqu...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com