Breeding method of strain with high yield of phenazine-1-formamide based on genome shuffling
A technology of genome shuffling and high-yield strains, applied in the field of bioengineering, can solve the problems of phenazine-1-carboxamide high-yield strain breeding that have not yet been reported, difficult work, chromosome damage, etc., to save labor costs and improve efficiency , the effect of simple operation
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
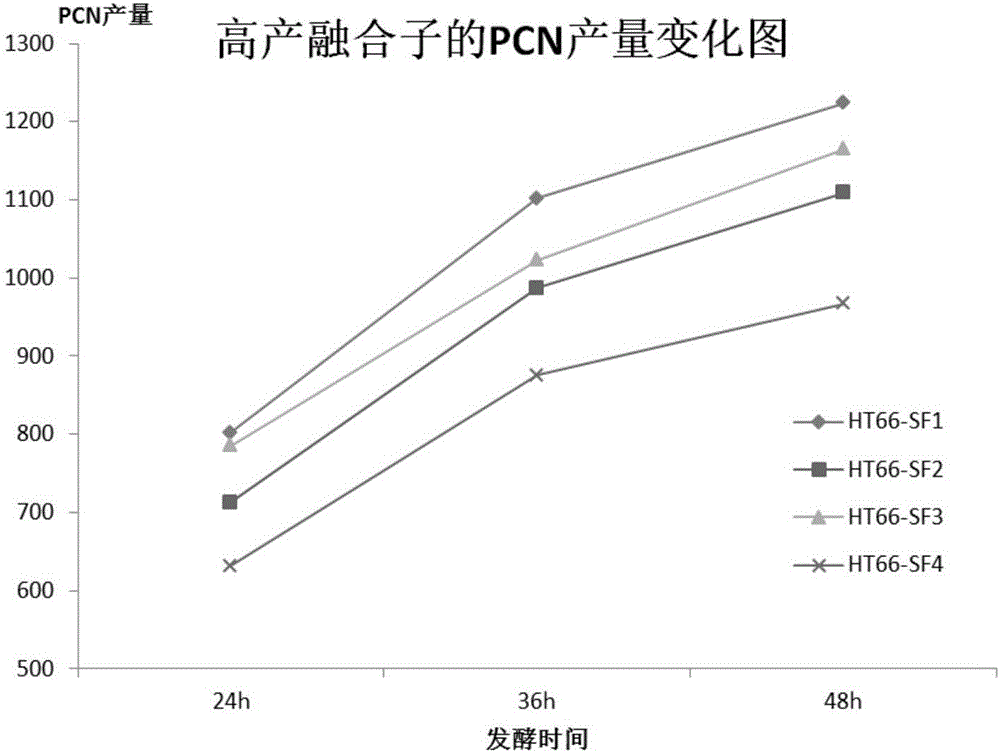
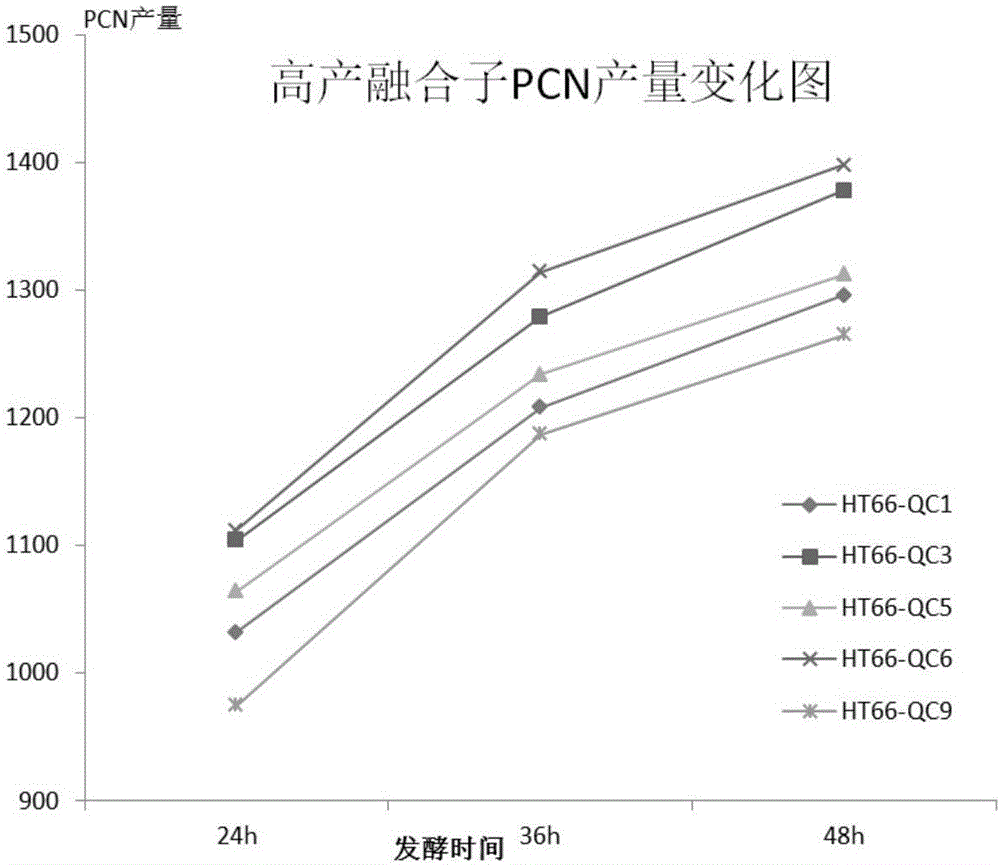
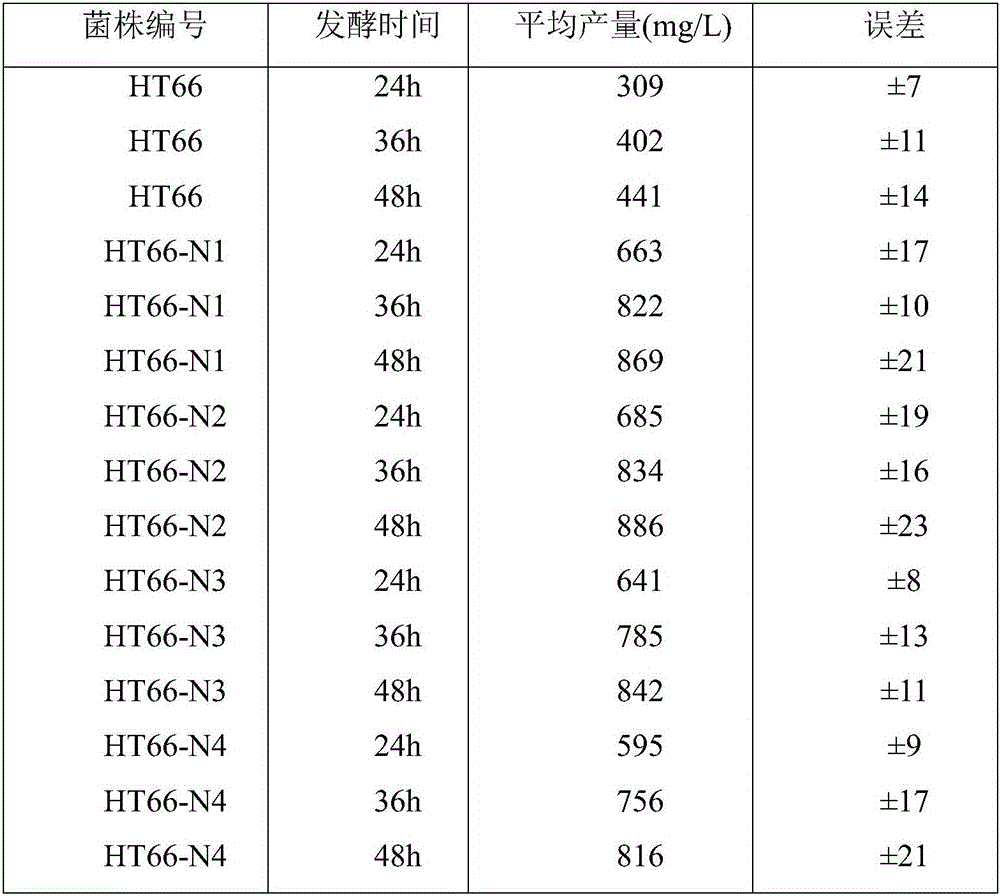
Image
Examples
Embodiment 1
[0034] 1.1 NTG mutagenesis and primary screening
[0035] In this example, Pseudomonas chloropinus HT66 was used as the starting strain, and a high-yield mutant strain was obtained by treatment with a chemical mutagen. The mutagenesis treatment may be NTG mutagenesis, but is not limited to this method. When using NTG mutagenesis treatment, the steps are as follows:
[0036]Prepare a NTG mother solution with a concentration of 10g / L and dilute it proportionally when used. Aspirate 1mL logarithmic phase bacterial suspension, wash the bacterial cells twice with sterile 0.01mol / L PBS buffer (pH7.4); The concentration was 50, 100, 200, 500, 1000g / L, and treated at 28°C for 20min; adding 0.5mL of 1mol / L NaCl solution to terminate the reaction, using sterile medium to wash the bacteria for 3 times to completely remove residual NTG, diluting appropriately, and then applying After 24 hours, count the number of colonies grown on the plate to obtain the lethal rate. When the dose o...
Embodiment 2
[0078] 2.1 Cobalt-60γ-ray mutagenesis and primary screening
[0079] In this example, Pseudomonas chloropinus HT66 was used as the starting strain, and a high-yield mutant strain was obtained by physical mutagenesis. The mutagenesis treatment may be cobalt-60γ-ray mutagenesis, but is not limited to this method. The specific method includes the following steps:
[0080] 1. Take out the hair strain HT66, activate and cultivate it as seed solution.
[0081] 2. Take the seed solution and centrifuge, suck off the culture medium, and resuspend the bacteria with PBS buffer to make the cell concentration about 10 7 to 10 8 individual / mL.
[0082] 3. Divide the cell suspension into sterilized glass sampling vials, and place them in a cobalt-60 gamma ray radiation environment to receive irradiation; the vials of the control bacteria solution do not receive irradiation.
[0083] The absorbed doses received by the cell suspensions in each group were 0.225, 0.320, 0.430, 0.490 and 0...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com