A Nucleic Acid Detection Technology Based on Multi-cross Isothermal Amplification Combined with Gold Nanobiosensor
A biosensor, gold nanotechnology, applied in the field of microbiology and molecular biology, which can solve the problems of error-prone, expensive, time-consuming and labor-intensive
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0062] Embodiment 1 Standard MCDA-LFB detects
[0063] 1. Standard MCDA reaction system
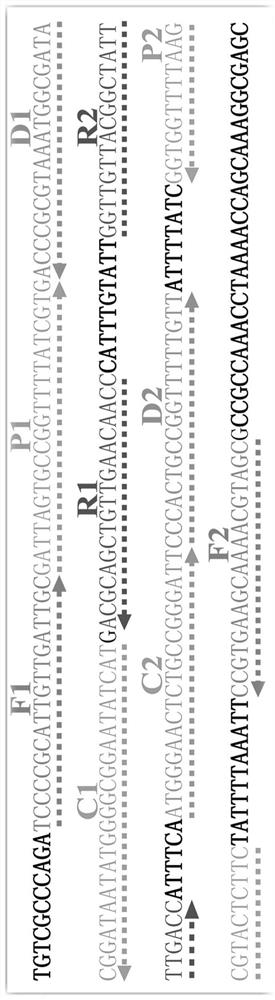
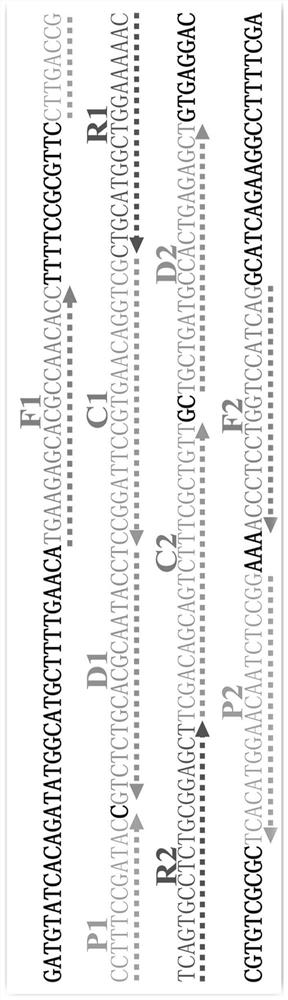
[0064] The concentration of cross primers CP1 and CP2 was 60 pmol, the concentration of displacement primers F1 and F2 was 10 pmol, the concentration of amplification primers R1, R2, D1 (D1*) and D2 (D2*) was 30 pmol, and the concentration of amplification primer C1 (C1* ) and C2 (C2*) at a concentration of 20pmol, Betain of 10mM, MgSO of 6mM 4 , 1 mM dNTP, 12.5 μL of 10×Bst DNA polymerase buffer, 10 U of strand-displacing DNA polymerase, 1 μL of template, and add deionized water to 25 μL. The entire reaction was kept at 65°C for 1 hour, and the reaction was terminated at 80°C for 5 minutes.
[0065] 2. Result detection
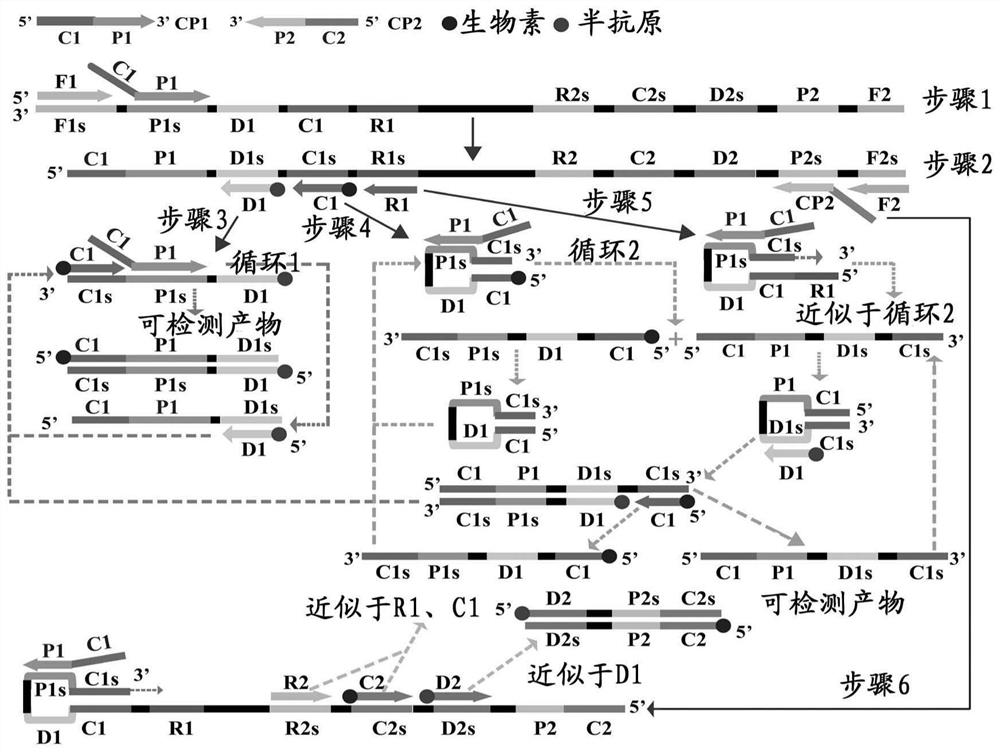
[0066] (1) Visible color change method: MCDA produces a large amount of pyrophosphate ions while synthesizing DNA, which can capture the manganese ions combined with calcein, so that calcein returns to a free state and fluoresces. The luminescent mixture can combine ...
Embodiment 2
[0075] Embodiment 2 multiple ET-MCDA reaction system:
[0076] ET-MCDA, that is, endonuclease-mediated real-time multiple cross-displacement nucleic acid amplification technology, see CN105755134A for details, and the present invention takes this patent document as a part of the disclosure content of the present invention by citing it.
[0077] The concentration of cross primers Sal-CP1 and Sal-E-CP1 is 30pmol, the concentration of Sal-CP2 is 60pmol, the concentration of displacement primers Sal-F1 and Sal-F2 is 10pmol, and the concentration of amplification primers Sal-R1, Sal-R2, Sal -The concentration of D1 and Sal-D2 is 30pmol, the concentration of amplification primers Sal-C1 and Sal-C2 is 20pmol, the concentration of cross primer Shi-CP1 and Shi-E-CP1 is 8pmol, and the concentration of Shi-CP2 is 16pmol, The concentration of displacement primers Shi-F1 and Shi-F2 is 10pmol, the concentration of amplification primers Shi-R1, Shi-R2, Shi-D1 and Shi-D2 is 8pmol, and the con...
Embodiment 3
[0079] Embodiment 3 multiple MCDA reaction system:
[0080] 1. Multiple MCDA reaction system
[0081] The concentration of cross primer Sal-CP1 and Sal-CP2 is 60pmol, the concentration of displacement primer Sal-F1 and Sal-F2 is 10pmol, and the concentration of amplification primer Sal-R1, Sal-R2, Sal-D1* and Sal-D2 is 30pmol, the concentration of amplification primers Sal-C1* and Sal-C2 is 20pmol, the concentration of cross primer Shi-CP1 and Shi-CP2 is 16pmol, the concentration of displacement primer Shi-F1 and Shi-F2 is 10pmol, the concentration of amplification primer Shi -The concentration of R1, Shi-R2, Shi-D1 and Shi-D2* is 8pmol, the concentration of amplification primers Shi-C1 and Shi-C2* is 10pmol, 10mM Betain, 6mM MgSO 4 , 1 mM dNTP, 12.5 μL of 10×Bst DNA polymerase buffer, 10 U of strand displacement DNA polymerase, 1 μL of templates for Salmonella and Shigella, and added deionized water to 25 μL. The entire reaction was kept at 65°C for 1 hour, and the reaction...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com