Method for detecting noninvasive paternity tests through high-throughput sequencing
A paternity testing, high-throughput technology, applied in the field of high-throughput sequencing detection non-invasive paternity testing, can solve problems such as immeasurable development prospects, and achieve the effect of low cost, accurate results and high throughput
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
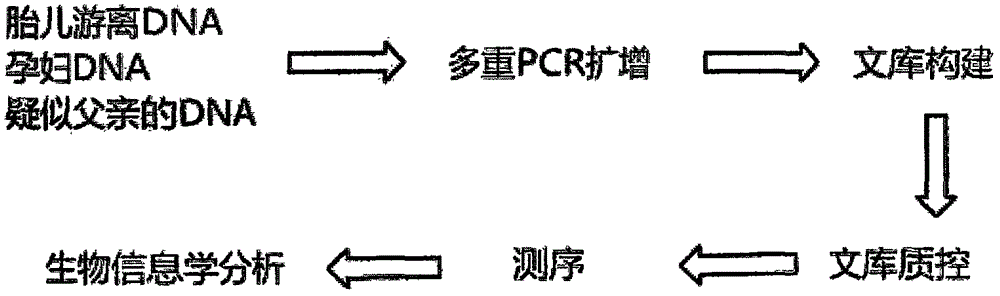
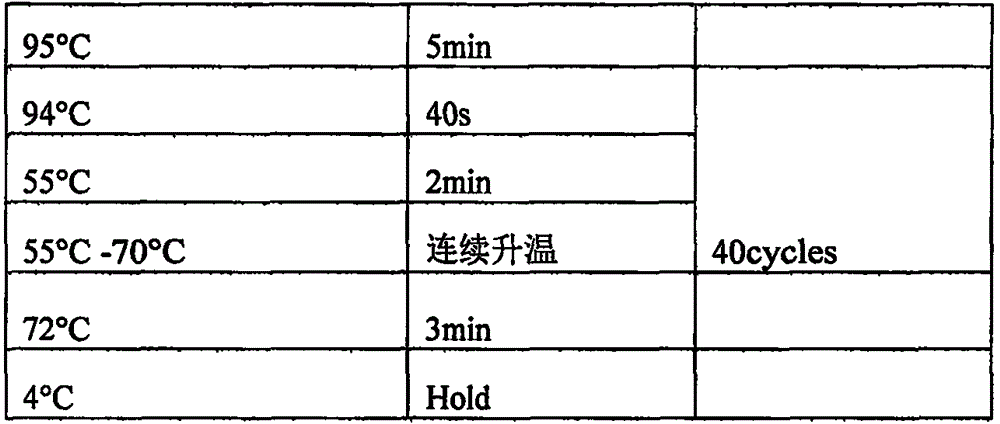
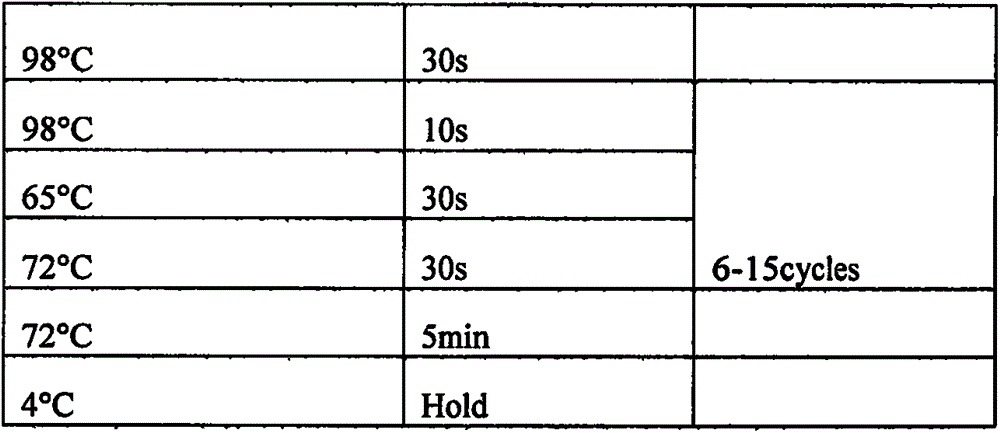
[0023] A method for high-throughput sequencing detection of non-invasive paternity testing, comprising the following steps:
[0024] Step a, extracting genomic DNA;
[0025] (1) Extract fetal cell-free DNA
[0026] 1) Take out 600μl plasma sample and place it at room temperature. After the sample melts, add 10μl proteinase K immediately, shake and mix, and centrifuge briefly;
[0027] 2) Immediately add 600μl GB Mix, shake and mix, and centrifuge briefly; after 10 minutes in a 56°C water bath, take out the sample tube, dry the tube wall, leave it at room temperature for 5 minutes, and centrifuge; put the adsorption column and collection tube together, and Sample information Mark the sample number on the tube cap;
[0028] 3) Add 300 μl of -20°C pre-cooled absolute ethanol, gently invert and mix 5-7 times, let stand for 5 minutes, and centrifuge;
[0029] 4) Transfer 750 μl of the sample solution to the adsorption column tube, centrifuge at 8000 rpm for 30 seconds, discard t...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com