Kit for quickly detecting Brettanomyces bruxellensis
A detection kit and technology of Brettanomyces, applied in the determination/inspection of microorganisms, biochemical equipment and methods, etc., can solve the problems of false negatives, no fast detection time for wine, etc., and achieve the effect of improving the current detection status
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
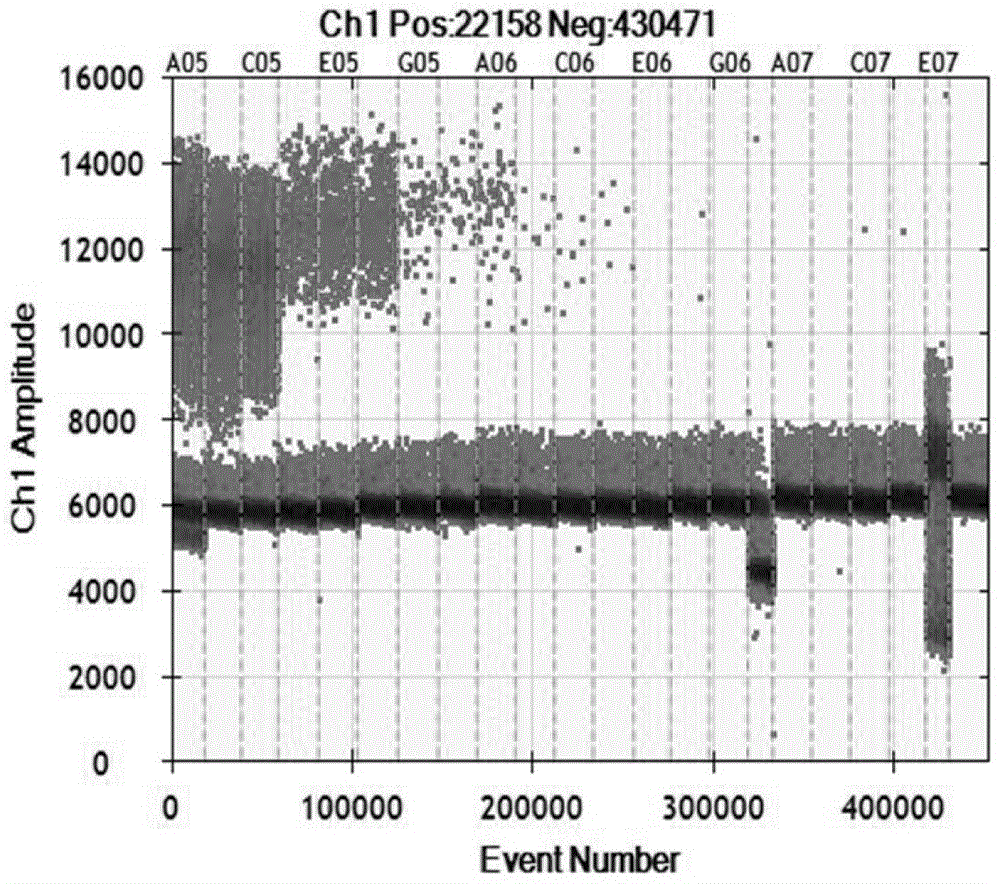
Image
Examples
Embodiment 1
[0018] Example 1: Extraction of DNA in Wine Samples
[0019] 1. Take 45 mL of the wine sample to be analyzed, place it in a 50 mL test tube with a sterile screw cap, centrifuge at 4 °C for 5 min with a centrifugal force of 9300×g, and discard the supernatant. Then add 45 mL of 10 mmol / L Tris-HCl pH 8.0 to wash the residue, vortex the above solution, centrifuge at 4 °C for 5 min with a centrifugal force of 9300×g, and discard the supernatant. Vortex gently to resuspend the pellet in the residual liquid.
[0020] 2. DNA extraction
[0022] Add 0.3 g of glass beads to the collected precipitate suspension, followed by PVPP to make a final mass / volume ratio of 1%. Add 200 µL of solution I and 200 µL of extract. The above solution was vortexed for 80 s, and then cooled at -20 °C for 80 s; the above vortex cooling step was repeated 3 times.
[0023] 2.2 DNA purification
[0024] 1) Add 200 μL TE solution, centrifuge at 12000×g for 5 min. Carefully colle...
Embodiment 2
[0031] Embodiment 2: digital PCR reaction
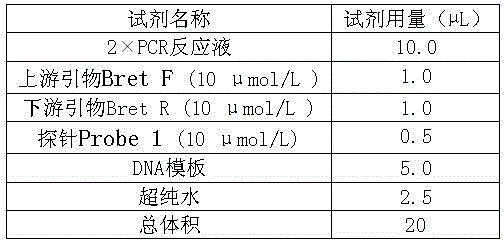
[0032] 1. PCR reaction system, see Table 1.
[0033] Table 1 Digital PCR reaction system
[0034]
[0035] 2. PCR reaction parameters: 95°C pre-denaturation for 10 min; 95°C denaturation for 30 sec, 55°C annealing extension for 30 sec, 72°C for 30 sec, 50 cycles; heating rate 2°C / min. Store the reaction product at 4°C.
Embodiment 3
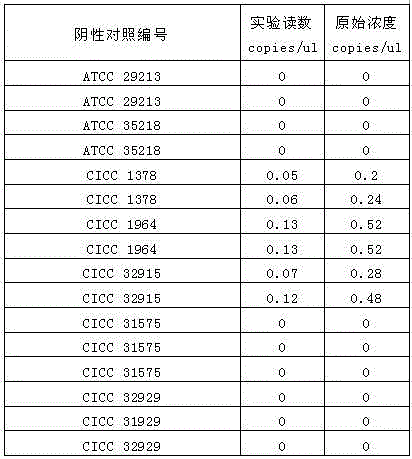
[0036] Embodiment 3: the sensitivity experiment of method
[0037] Mock samples: Wine samples were tested with a kit for the rapid detection of Brettanomyces brussels in samples. The kit consists of:
[0038] Reagent A: wine sample DNA extraction reagent;
[0039] Reagent B: PCR reaction solution, which contains the upstream primer Bret F, its sequence is: 5'-GTTCACACAATCCCCTCGATCAAC-3';
[0040] Reagent C: PCR reaction solution, which contains the downstream primer Bret R, whose sequence is:
[0041] 5'-GTTCACACAATCCCCTCGATCAAC-3';
[0042] Reagent D: PCR reaction solution, which contains a probe whose sequence is: Probe 1 5'-FAM-ATGGCGAGGATGAAAGTTTGGGATACA-BHQ1-3';
[0043] Reagent E: 2×PCR reaction solution (without dUTP).
[0044] First use reagent A to extract the DNA in the sample, then perform PCR reaction, and perform data analysis after the reaction to quantify Brettanomyces brussels in the sample.
[0045] After diluting the Brettanomyces brussels standard stra...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com