Method and device for detecting designated location point of variation
A mutation and site technology, applied in the device for detecting fusion gene mutation, in the field of fixed-point mutation detection, can solve the problems of increased workload, difficulty in detection, lack of effective guidance, etc., and achieve the effect of accurate and efficient detection
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
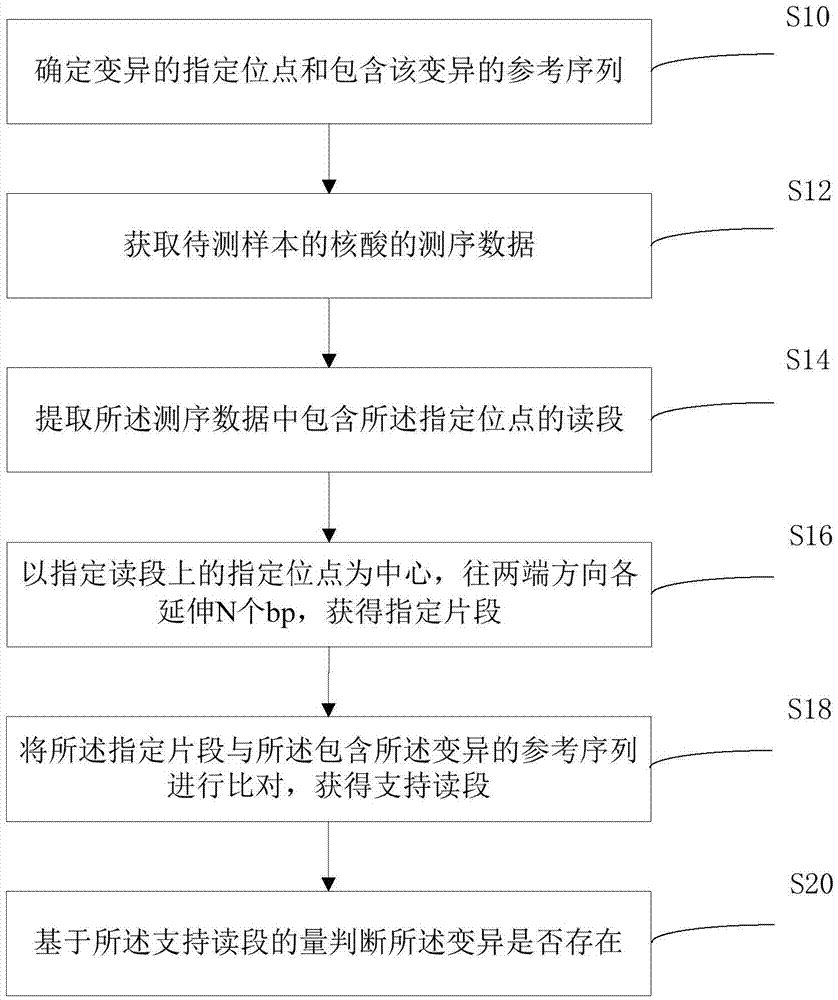
Embodiment 1
[0063] (1) Construction of reference value model
[0064] 1. Hypothetical basis for the construction of the reference value model
[0065] 1.1. For any site, assume that the base corresponding to the reference genome is r∈{A,T,C,G};
[0066] 1.2, for any site, assume that the corresponding base of all reads covering the site is b i , the base quality value is q i , the corresponding base error rate is i=1,2,...,d, where d represents the sequencing depth corresponding to this site.
[0067] 2. Model establishment
[0068] The data distribution of each site is divided into two models to explain:
[0069] Model M 0 : There is no variation at this site, and those bases that are different from the reference genome are caused by systematic errors;
[0070] Model The mutation r→m at this site really exists, and the allelic mutation frequency is f, and those bases that are neither r nor m are treated as systematic errors.
[0071] The data distribution of this site can be...
Embodiment 2
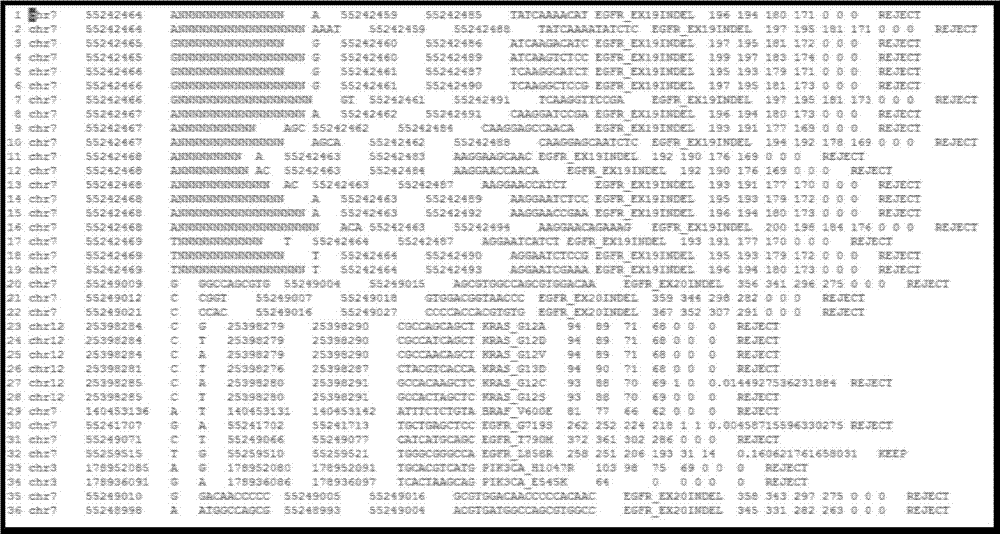
[0096] After obtaining the off-machine sequencing data, taking the off-machine data from the BGISEQ-100 platform as an example, the mutation detection generally includes the following parts:
[0097] 1. Variation known information processing and sequencing data preprocessing
[0098] 1.1 Convert the type of variation to be detected into the format recognized by the detection program, and generate a list of variation to be detected.
[0099] 1.2 Compare the off-machine data with the reference genome. The effective sequencing data of BGISEQ-100 was aligned to the reference genome using the tmap tool to obtain accurate alignment results. The tmap tool comes from: https: / / github.com / iontorrent / TS / tree / master / Analysis / TMAP.
[0100] Sort. Use samtools sort to sort the results (bam files) after using tmap comparison: sort according to the chromosome number and the position on the chromosome in ascending order.
[0101] The PCR duplicate fragments of the alignment results were re...
Embodiment 3
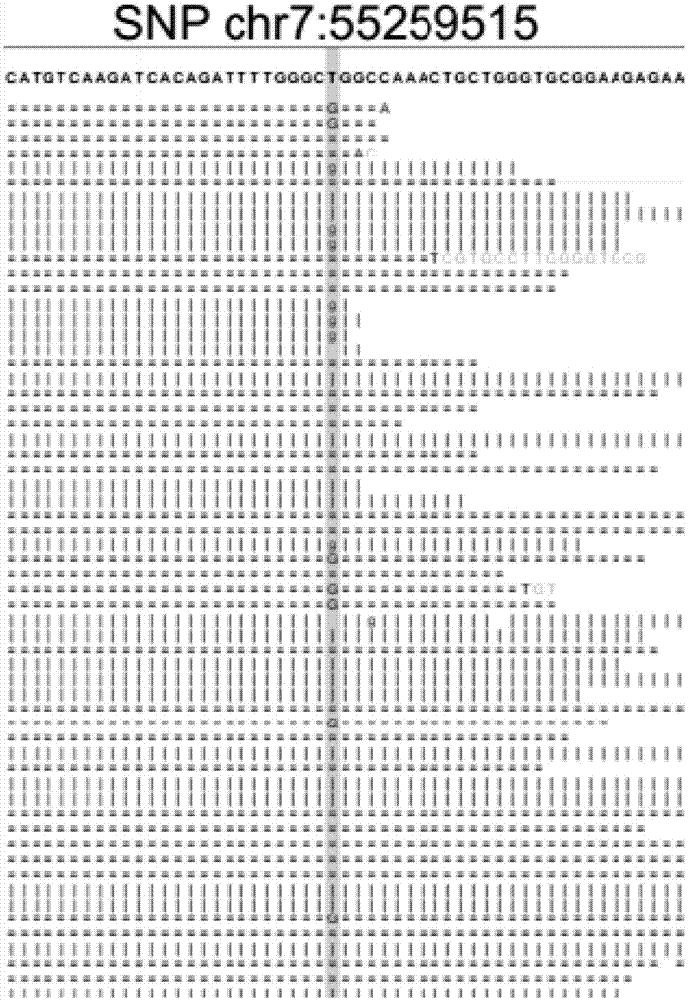
[0136] This example uses the FFPE tissue sample of a female patient with upper left lung adenocarcinoma to capture the target area and sequence it on the BGISEQ-100 platform. The effective data off the machine is compared by tmap, sorted by samtools, deduplicated by BamDuplicates, and samtools Index construction, mutation detection at known sites, variant annotation, report generation and other steps, finally obtain the patient's known site mutation detection report.
[0137] All the parts of the process of the above-mentioned variation detection method are integrated into the software Otype. The operating environment of the software is the Linux operating system. The specific operation steps are as follows:
[0138] Enter the following command line in the Linux operating system computer terminal:
[0139] perlOtype.pl–lsample.list–o outdir–O run.sh will generate the corresponding running script.
[0140] sh run.sh runs the script.
[0141] For the meaning of the command lin...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com