A screening system and application of regγ-20s proteasome inhibitors
A proteasome inhibitor, inhibitor technology, applied in the field of biochemistry and biomaterials, to achieve the effect of high sensitivity, high specificity, and efficient detection ability
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
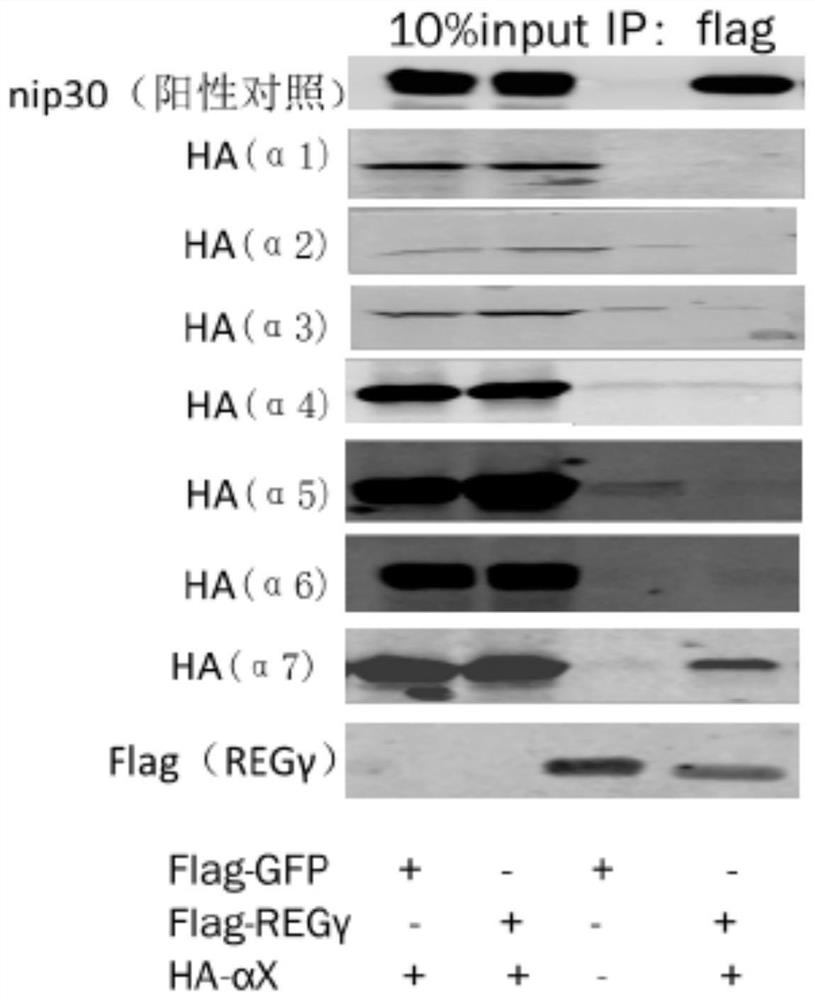
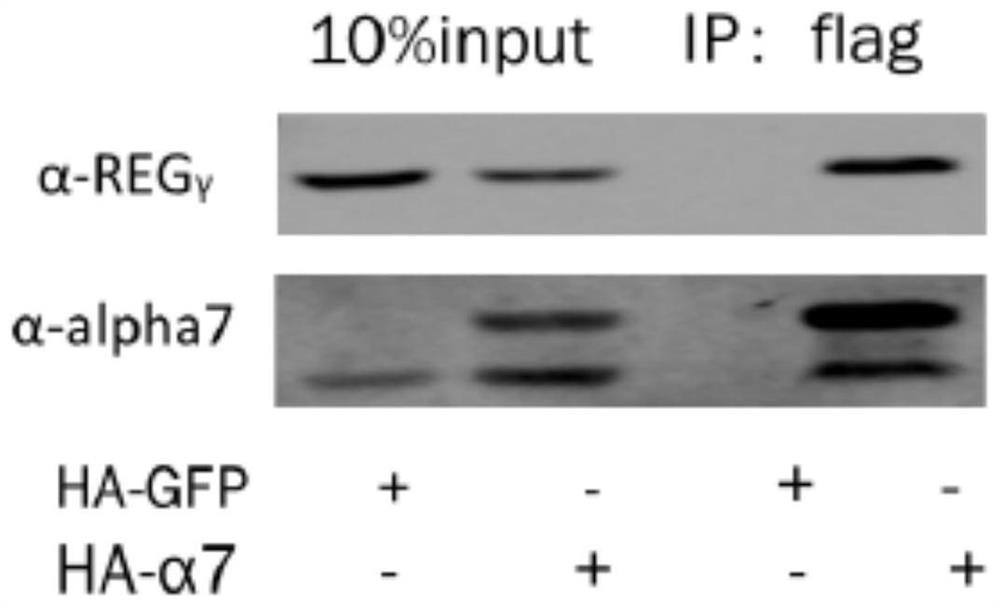
[0038] Example 1 Co-immunoprecipitation experiment proves that REGγ specifically binds to α7 subunit of 20S proteasome
[0039] The α subunit of the 20S proteasome is its regulatory subunit, which is opened or closed under the control of activating factors. REGγ can promote the degradation of substrate proteins. It is likely that REGγ may open the gate of 20S by combining with the α subunit. The present invention The interaction between REGγ and 20S proteasome α1-α7 was detected by co-immunoprecipitation.
[0040] Specific experimental methods:
[0041] 1. Before transfection, HeLa cells were planted in six-well plates at a concentration of 60%, and cultured for 12 hours.
[0042] 2. Add 300 μL of serum-free DMEM medium to a 1.5 mL centrifuge tube, and then add the target plasmid and Young’s transfection reagent. The ratio of the two is 1 μg: 2 μL. Each well of a six-well plate can transfer 1 μg of plasmid.
[0043] 3. Vortex and vibrate for 10 seconds to mix well. After sta...
Embodiment 2
[0052] Example 2 Yeast two-hybrid experiment proves that REGγ specifically binds to α7 subunit of 20S proteasome
[0053] Using different experiments, it was again demonstrated that REGγ specifically binds to the α7 subunit of the 20S proteasome.
[0054] PSMA1-PSMA7 (α 1 -α 7 ) was constructed on the pGADT7 vector, and the yeast two-hybrid experiment was used to further verify the interaction between REGγ and α7.
[0055] The results of yeast two-hybrid image 3 As shown, only when PGAD-α7 / pGBK-REGγ was co-transfected, yeast could grow on the four-deficient plate, that is to say, REGγ only interacted with the α7 subunit, which was consistent with the results of co-immunoprecipitation ( nip30 as a positive control).
[0056] The specific experimental method of yeast two-hybrid experiment:
[0057] 1. Take out the yeast strain from the -80°C refrigerator and melt it at room temperature, then burn the inoculation loop on the alcohol lamp until it is red hot, and after cooli...
Embodiment 3
[0068] Example 3 Using luciferase fragment complementation technology to construct a screening system for inhibitors of the interaction between REGγ and α7
[0069] By comparing several commonly used molecular imaging techniques for studying protein-protein interactions, such as energy resonance transfer technology, green fluorescent protein fragment complementation technology and luciferase fragment complementation technology, etc. The present invention finally selects the highly efficient and convenient luciferase fragment complementation technology as the system basis for screening the inhibitors of the interaction between REGγ and α7. In the design experiment, the present invention considers three key issues: the first key issue is (1) the cleavage site of luciferase, how to cut the luciferase so that the two fragments produced can recombine well , and the newly bound luciferase can regain luciferase activity. Such as Figure 4 As shown, the present invention selects the...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com