Analysis method of genomic DNA (deoxyribonucleic acid) fragment editing precision applicable to CRISPR/Cas9 (clustered regularly interspaced short palindromic repeats/CRISPR-associated nuclease 9) system, and application
An analysis method and genome technology, applied in the field of analysis of the accuracy of genome DNA fragment editing, to achieve the effect of improving experimental efficiency
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
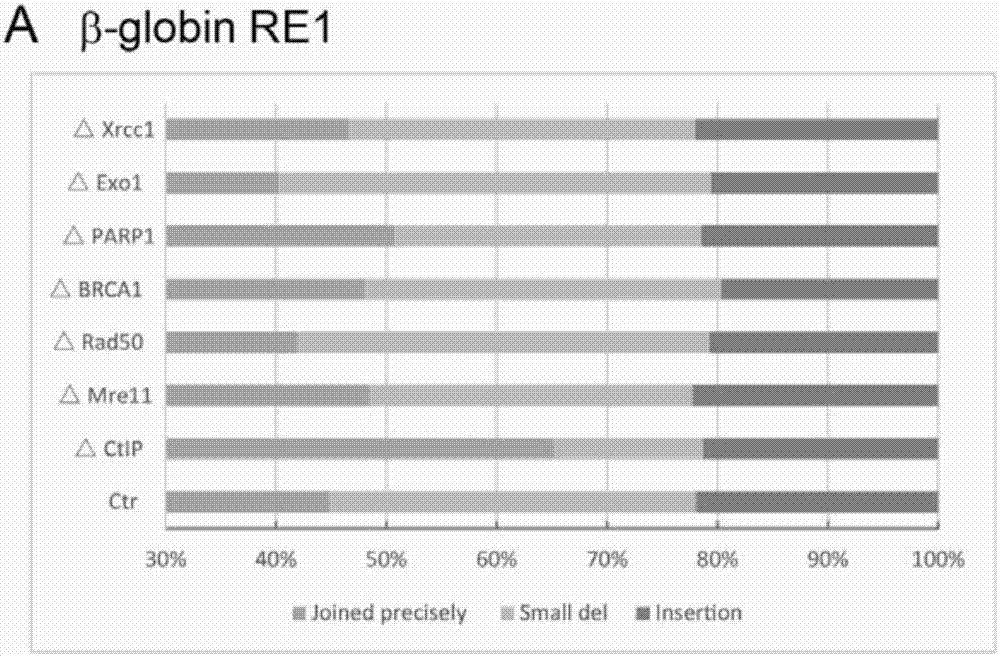
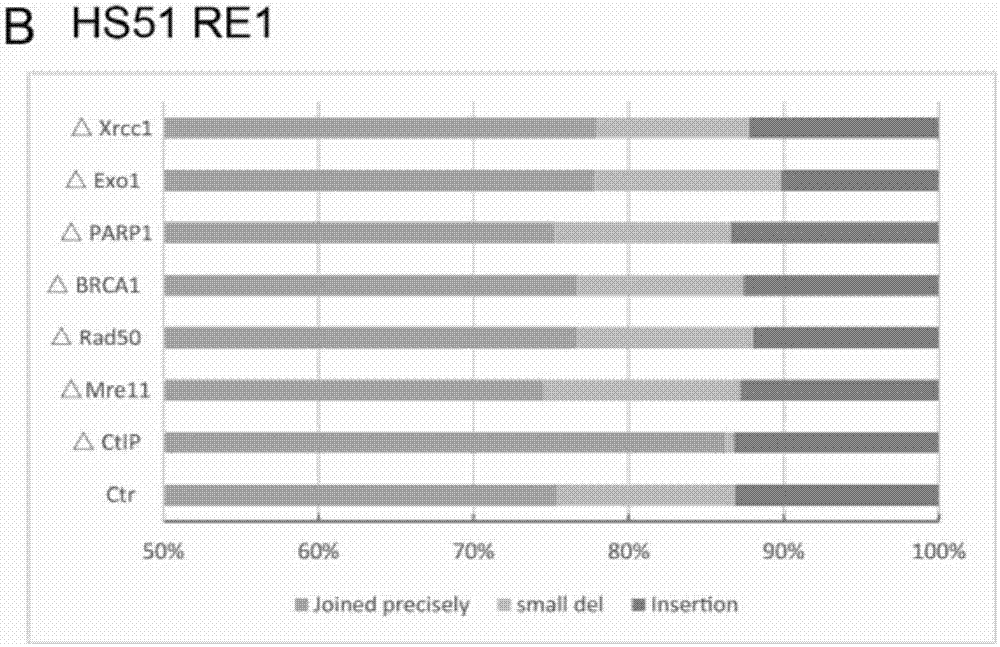
[0186] Example 1 Transfection of sgRNAs targeting the CtIP gene can improve the precise ligation efficiency after deletion of DNA fragments
[0187] 1. Construction of sgRNAs plasmids for STM sites and CtIP genes
[0188] (1) Purchase primers
[0189] Purchase forward and reverse deoxyoligos with 5' hanging ends "ACCG" and "AAAC" that can be complementary paired for the STM site (β-globin RE1) and the sgRNAs targeting sequence of the CtIP gene from Shanghai Sunny Biotechnology Co., Ltd. Nucleotides.
[0190] Forward and reverse deoxy oligonucleotides:
[0191] β-globin RE1sgRNA1F: accgATTGTTGTTGCCTTGGAGTG (SEQ ID NO.1)
[0192] β-globin RE1sgRNA1R: aaacCACTCCAAGGCAACAACAAT (SEQ ID NO.2)
[0193] β-globin RE1sgRNA2F: accgCTGGTCCCCTGGTAACCTGG (SEQ ID NO.3)
[0194] β-globin RE1sgRNA2R: aaacCCAGGTTACCAGGGGACCAG (SEQ ID NO.4)
[0195] CtIPsgRNA1F: accgGAGCAGAGCAGCGGGGCAA (SEQ ID NO.5)
[0196] CtIPsgRNA1R: aaacTTGCCCCGCTGCTCTGCTC (SEQ ID NO. 6)
[0197] CtIPsgRNA2F: accgTT...
Embodiment 2
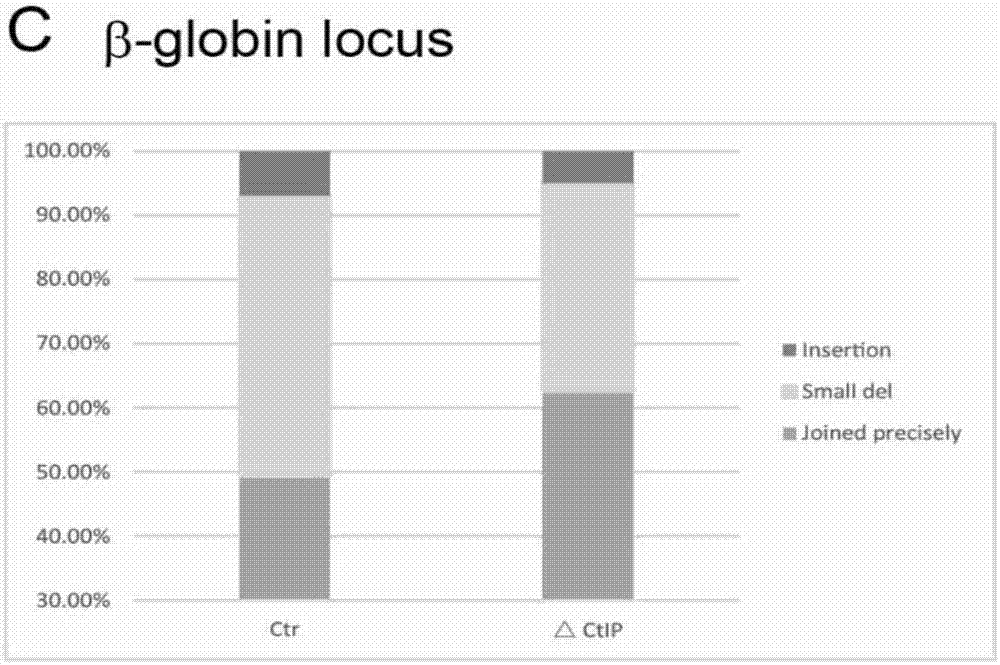
[0267] Example 2 CtIP mutations in cell lines can effectively improve the precision ligation efficiency of deletion of target DNA fragments
[0268] 1. Obtain CtIP-mutated cell lines by CRISPR system
[0269] 1) HEK293T cells were cultured in a culture flask, and when they grew to 80-90% of the culture flask, the grown cells were plated in a 12-well plate with DMEM complete antibiotic-free medium, and cultured overnight. When the cells in the 12-well plate grew to 80-90%, the prepared humanized Cas9 plasmid (800ng) and the sgRNAs plasmid (600ng each) at the CtIP site were transfected with Lipofectamine2000.
[0270] 2) Add Puromycin (2μg / ml) to the cells 48 hours after transfection for four days of drug screening, then culture them in fresh medium for eight days, collect the cells, count the cells evenly dispersed, and then dilute to a certain number of species Into a 96-well plate (only one cell per well), after 6 days of culture, the well plate with only one cell cluster co...
Embodiment 3
[0287] Example 3 3-AP Improves the Precision Ligation Efficiency of DNA Fragment Deletion
[0288] 1. Cell line transfection with Lipofectamine 2000 at the STM site
[0289] HEK293T cells and CtIP mutant cells were plated in 12-well plates with DMEM complete anti-antibody medium and cultured overnight. When the cells in the 12-well plate grew to 80-90%, the medium was removed, and DMEM containing DMSO or different concentrations of 0.2 μM, 0.4 μM, 0.8 μM, and 1.6 μM 3-AP (SML0568, Sigma) was added without Anti-medium, the prepared humanized Cas9 plasmid (800ng) and sgRNAs (600ng each) targeting the STM site were transfected with Lipofectamine 2000. After 24 hours, remove the medium, add DMEM complete double antibody medium (add 10% fetal bovine serum and 1% penicillin double antibody), and after another 24 hours, collect the cells, use the genome extraction kit ( Genomic DNA Purificationkit, Promega) was used to extract the genome, and each sample had two replicates.
[0...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com