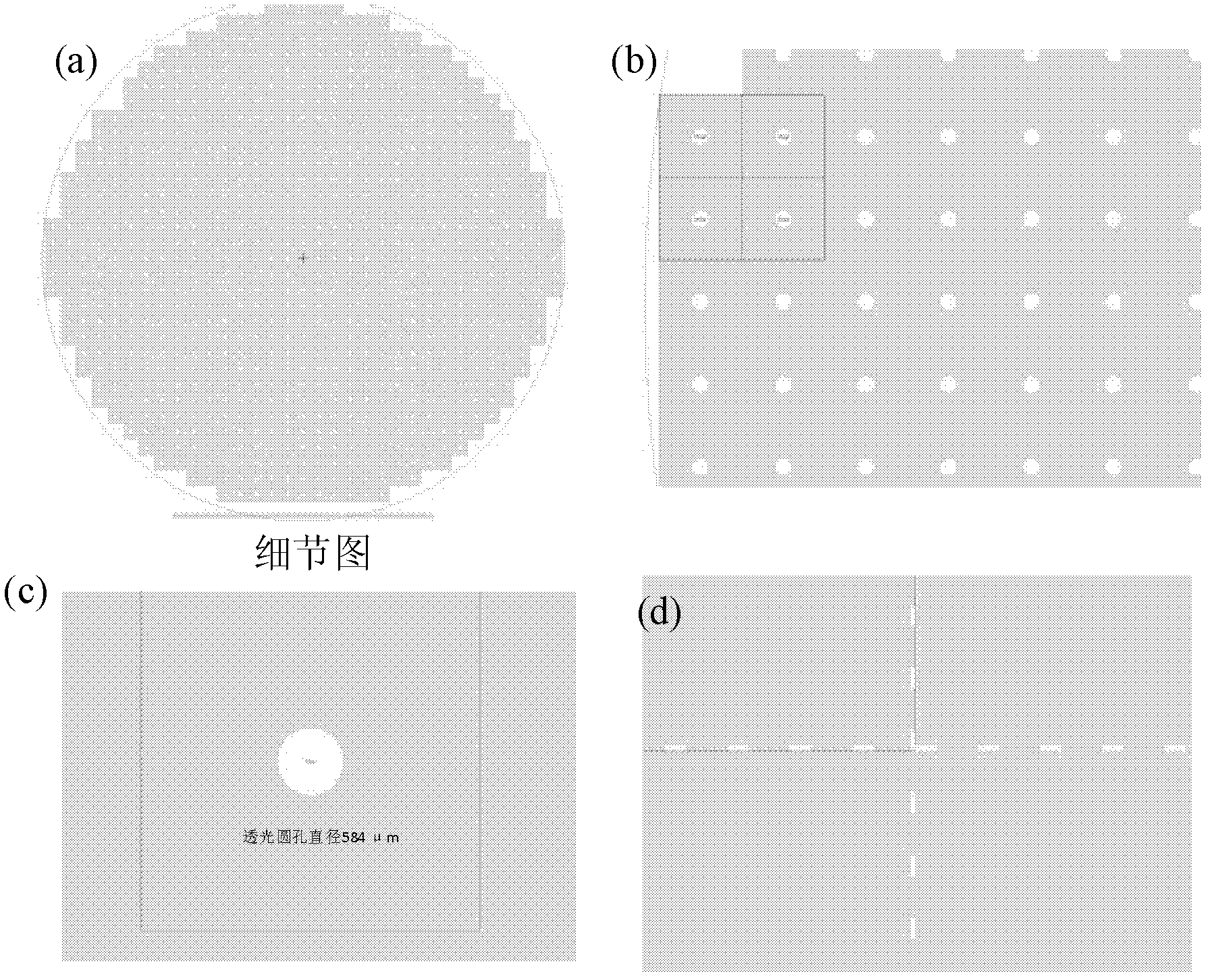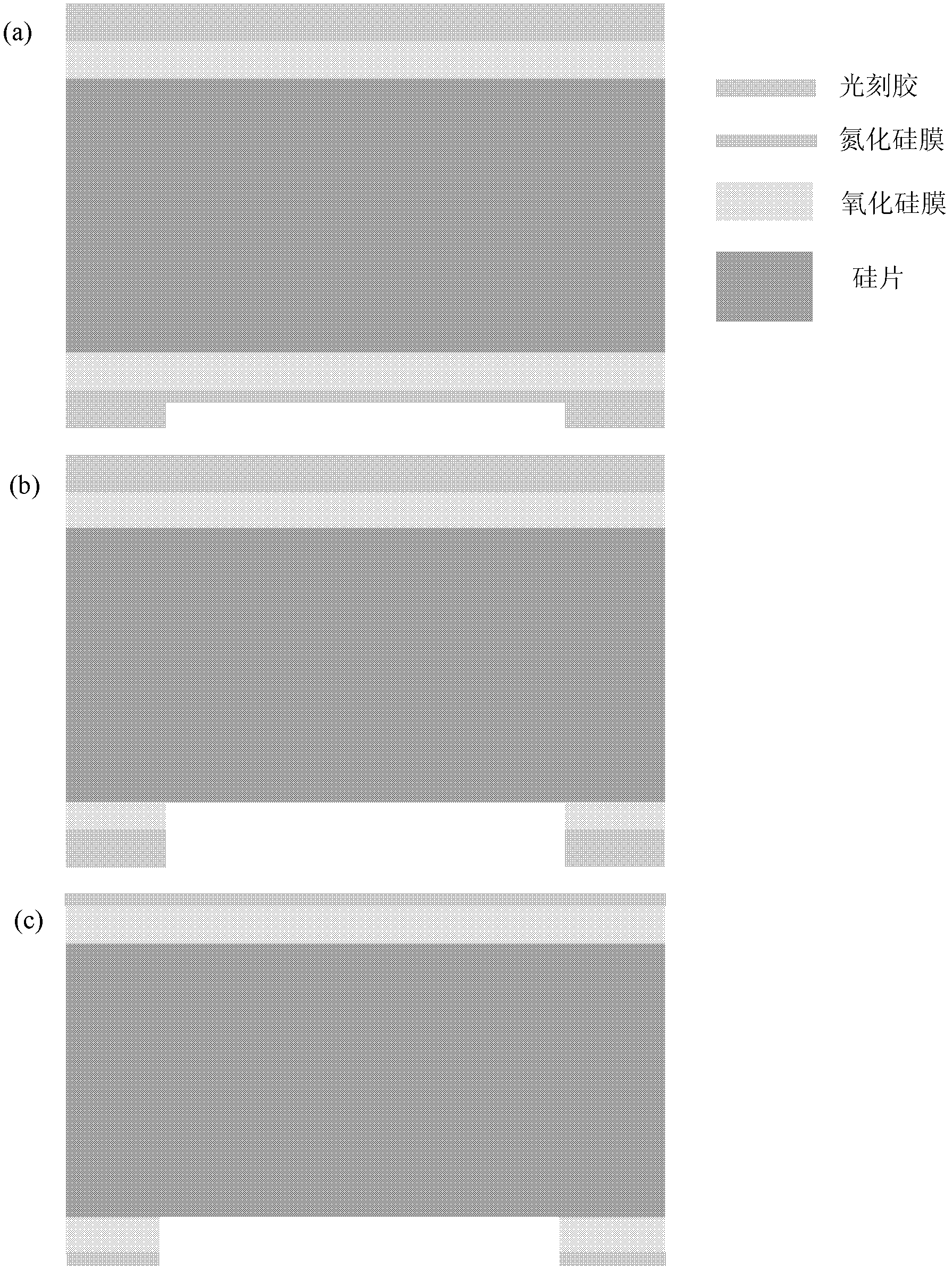Method for decelerating nucleic acid molecules in solid nanopore
A nucleic acid molecule and nanopore sequencing technology, which is applied in biochemical equipment and methods, analytical materials, and the determination/inspection of microorganisms. and improved controllability, high current signal-to-noise ratio, and improved time resolution
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment
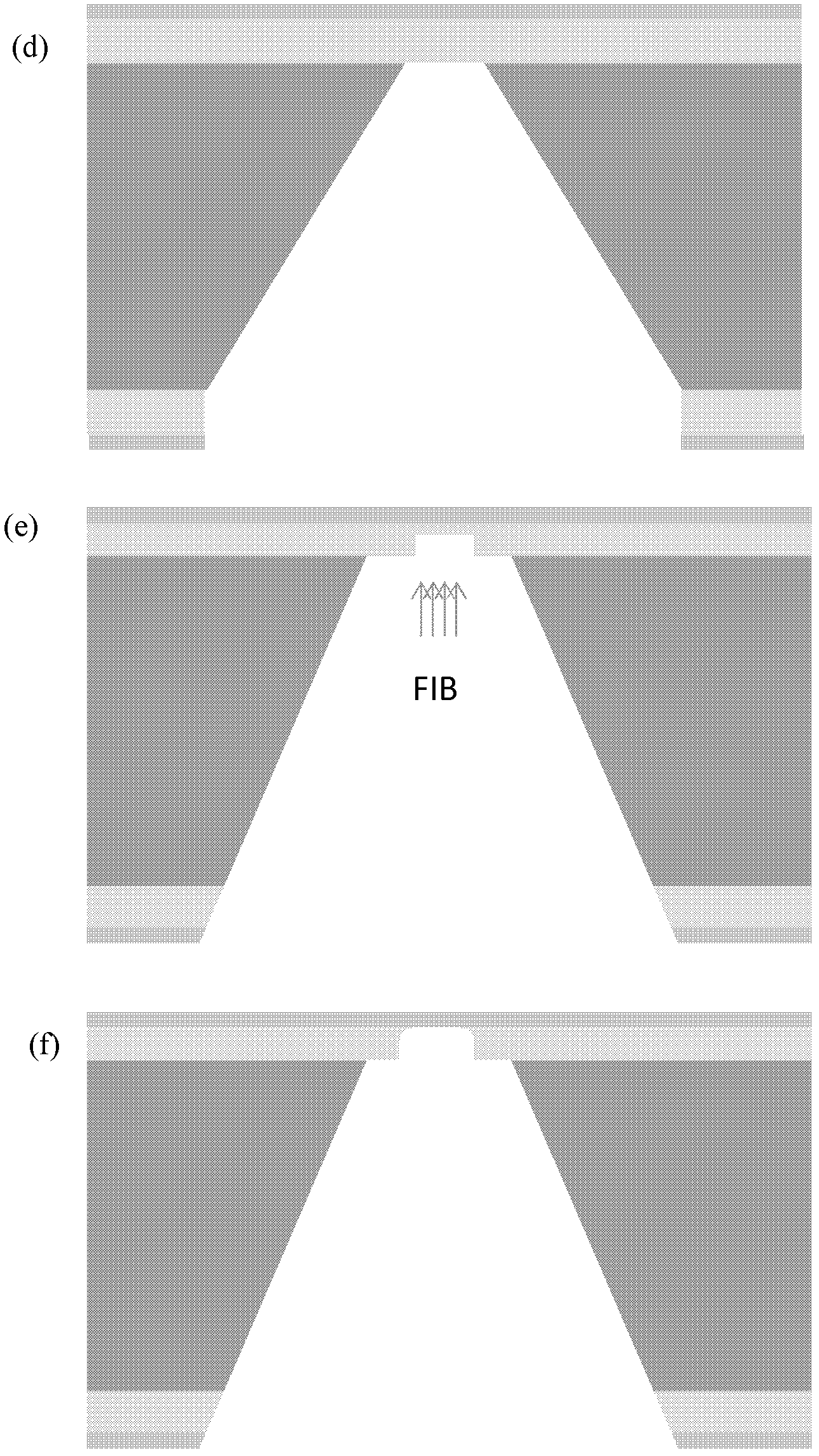
[0027] 1) Preparation of chip devices
[0028] The purpose of this step is to prepare a solid-state nanoporous device that can be operated in a transmission electron microscope, so that the high-energy convergent electron beam in the transmission electron microscope can be used to punch holes in the suspended film of the device, thereby realizing a nanoporous device. This includes a series of micro-nano processing technology involved in traditional semiconductor processing technology, and also involves a series of modern cutting-edge nano-scale processing technology.
[0029] The specific method is as follows: First, a 4-inch silicon wafer with a (100) surface is grown with 2 microns of silicon oxide on both sides, and low-stress silicon nitride of about 150 to 200 nanometers is deposited by a low-pressure chemical vapor deposition method. Then make a photolithography mask, the purpose is to make a lot of 3mm×3mm small substrate periodic distribution on a 4-inch silicon wafer ...
PUM
| Property | Measurement | Unit |
|---|---|---|
| Diameter | aaaaa | aaaaa |
| Hole depth | aaaaa | aaaaa |
Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com